Lecture 10 - Property & Reaction Prediction#
Learning goals#
Set up Chemprop v2 for regression and classification on our C-H oxidation dataset.
Train four single task models for: Solubility, pKa, Melting Point, Toxicity.
Train a reactivity classifier and an atom-level selectivity predictor.
Interpret a trained model with Shapley values (SHAP) at the feature and node levels.
For this lecture 10, it is recommended to run everything in Colab. On this HTML page, some outputs are disabled due to execution limits.
1. Directed message-passing neural network (D-MPNN)#
We will train models for four molecular properties and reaction-related labels using Chemprop.
Briefly speaking, Chemprop builds neural models for molecules using a directed message passing neural network (D-MPNN).
As you recall from previous lecture, a message passing neural network (MPNN) updates hidden vectors on nodes and edges with local neighbor information, then an aggregation step creates a graph-level vector for prediction.
Chemprop’s directed variant changes the way messages flow: instead of passing information back and forth between atoms, it assigns a hidden state to each directed bond (atom i → atom j). This prevents immediate backtracking (“tottering”) where messages would simply bounce between two atoms without capturing new context. By using directed bonds, the model distinguishes subtle chemical environments. For example, the information carried from a carbon toward a nitrogen can be different than the reverse direction, which matters for reactivity and selectivity.
As a GNN, Chemprop also featurizes a molecule as a graph:
Nodes are atoms with features like atomic number, degree, aromaticity.
Edges are bonds with features like bond order and stereo.
Initial directed bond state \(h_{i→j}^{(0)}\) is a learned function of the source atom features and the bond features. For t = 1..T, update
\(
h_{i \to j}^{(t)} = \sigma \Big( W \cdot \big( h_{i \to j}^{(t-1)} + \sum_{k \in \mathcal{N}(i) \setminus \{j\}} h_{k \to i}^{(t-1)} \big) + b \Big)
\),
where σ is an activation such as ReLU, W is a learned weight, \(x_{i→j}\) are featurized inputs, \(⊕\) is concatenation. After T steps, Chemprop aggregates per directed bond states to atom states, then pools to a molecule vector \(h_mol\) using sum or mean or attention pooling. \(h_mol\) feeds a multitask feedforward head.
We have been working with the following quite many times:
Solubility_mol_per_L: continuous. Regression with loss like MSE or MAE.
pKa: continuous. Regression.
Melting Point: continuous. Regression.
Toxicity: categorical with values like
toxicornon_toxic. Binary classification.
While these two we never try before:
Reactivity: binary label
1vs-1. Binary classification. In our C-H oxidation dataset, this means whether the substrate will undergo oxidation.Site Selectivity: a set of atom indices. Atom-level classification inside a molecule. In our C-H oxidation dataset, this means which atom(s) are most likely to oxidize under certain electrochemical reaction condition, expressed as atom indices in the SMILES.
As a reminder, below are some reference formulas:
Regression losses
$\( \text{MSE} = \frac{1}{n}\sum_i (y_i - \hat y_i)^2,\qquad \text{MAE} = \frac{1}{n}\sum_i |y_i - \hat y_i| \)$Binary cross entropy
$\( \mathcal{L} = -\frac{1}{n}\sum_i \big(y_i\log \hat p_i + (1-y_i)\log(1-\hat p_i)\big) \)$
You saw this idea in earlier lectures. The new part is that Chemprop builds the graph from SMILES and offers modules for molecule, reaction and atom/bond tasks.
We begin with load and inspect the C-H oxidation dataset.
2. Data preparation#
url = "https://raw.githubusercontent.com/zzhenglab/ai4chem/main/book/_data/C_H_oxidation_dataset.csv"
df_raw = pd.read_csv(url)
df_raw.head(5)
| Compound Name | CAS | SMILES | Solubility_mol_per_L | pKa | Toxicity | Melting Point | Reactivity | Oxidation Site | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 3,4-dihydro-1H-isochromene | 493-05-0 | c1ccc2c(c1)CCOC2 | 0.103906 | 5.80 | non_toxic | 65.8 | 1 | 8,10 |
| 1 | 9H-fluorene | 86-73-7 | c1ccc2c(c1)Cc1ccccc1-2 | 0.010460 | 5.82 | toxic | 90.0 | 1 | 7 |
| 2 | 1,2,3,4-tetrahydronaphthalene | 119-64-2 | c1ccc2c(c1)CCCC2 | 0.020589 | 5.74 | toxic | 69.4 | 1 | 7,10 |
| 3 | ethylbenzene | 100-41-4 | CCc1ccccc1 | 0.048107 | 5.87 | non_toxic | 65.0 | 1 | 1,2 |
| 4 | cyclohexene | 110-83-8 | C1=CCCCC1 | 0.060688 | 5.66 | non_toxic | 96.4 | 1 | 3,6 |
# Clean a copy and normalize a few columns
df = df_raw.copy()
# Toxicity -> binary string 'toxic'/'non_toxic' to 1/0 if present
tox_map = {"toxic": 1, "non_toxic": 0}
if "Toxicity" in df:
df["tox_bin"] = df["Toxicity"].str.lower().map(tox_map)
# Reactivity -> 1/-1 to 1/0
if "Reactivity" in df:
df["react_bin"] = df["Reactivity"].map(lambda x: 1 if x==1 else 0)
# Oxidation Site -> list of ints
def parse_sites(x):
if isinstance(x, str) and len(x.strip())>0:
return [int(v) for v in x.split(",")]
return []
df["site_list"] = df["Oxidation Site"].apply(parse_sites)
# Take log of solubility (keep same column name)
if "Solubility_mol_per_L" in df:
df["logS"] = np.log10(df["Solubility_mol_per_L"] + 1e-6)
df[["SMILES","logS","pKa","Toxicity","Melting Point","react_bin","site_list"]].head(8)
| SMILES | logS | pKa | Toxicity | Melting Point | react_bin | site_list | |
|---|---|---|---|---|---|---|---|
| 0 | c1ccc2c(c1)CCOC2 | -0.983356 | 5.80 | non_toxic | 65.8 | 1 | [8, 10] |
| 1 | c1ccc2c(c1)Cc1ccccc1-2 | -1.980414 | 5.82 | toxic | 90.0 | 1 | [7] |
| 2 | c1ccc2c(c1)CCCC2 | -1.686343 | 5.74 | toxic | 69.4 | 1 | [7, 10] |
| 3 | CCc1ccccc1 | -1.317782 | 5.87 | non_toxic | 65.0 | 1 | [1, 2] |
| 4 | C1=CCCCC1 | -1.216890 | 5.66 | non_toxic | 96.4 | 1 | [3, 6] |
| 5 | C1CCSC1 | -0.917634 | 5.97 | non_toxic | 15.8 | 1 | [3, 5] |
| 6 | CN1CCCC1=O | -0.499442 | 5.91 | non_toxic | 71.1 | 1 | [3] |
| 7 | COCc1ccccc1 | -1.070756 | 5.61 | non_toxic | 108.5 | 1 | [3] |
⏰ Exercise 1
Count number of postive and negative reaction outcomes in react_bin.
We will create MoleculeDatapoint objects from SMILES and targets, split the data, and build loaders.
Our first target will be solubility.
Step 1. Build datapoints.
Each row of the dataframe is now represented as a MoleculeDatapoint. It stores the SMILES, the numeric target (solubility here), plus metadata like optional weights.
This is the atomic unit Chemprop will pass to the featurizer.
# Keep rows that have both SMILES and solubility
df_sol = df[["SMILES","logS"]].dropna()
smis = df_sol["SMILES"].tolist()
ys = df_sol["logS"].to_numpy().reshape(-1,1)
sol_datapoints = [data.MoleculeDatapoint.from_smi(smi, y) for smi, y in zip(smis, ys)]
len(sol_datapoints), sol_datapoints[0].y.shape
(575, (1,))
sol_datapoints[0]
MoleculeDatapoint(mol=<rdkit.Chem.rdchem.Mol object at 0x797c8982cdd0>, y=array([-0.9833561]), weight=1.0, gt_mask=None, lt_mask=None, x_d=None, x_phase=None, name='c1ccc2c(c1)CCOC2', V_f=None, E_f=None, V_d=None)
Step 2. Split to train, val, test.
We divided the list of datapoints into three folds.
Training: used to fit model weights.
Validation: used to monitor progress and stop early.
Test: kept blind until the end.
Even though we used a random split here, Chemprop also supports scaffold-based splits which are often better for chemistry.
mols = [dp.mol for dp in sol_datapoints]
train_lists, val_lists, test_lists = make_split_indices(
mols=mols,
split="random",
sizes=(0.8, 0.1, 0.1),
seed=0,
num_replicates=1
)
train_dpss, val_dpss, test_dpss = split_data_by_indices(
sol_datapoints, train_lists, val_lists, test_lists
)
print(len(train_dpss[0]), len(val_dpss[0]), len(test_dpss[0]))
460 57 58
Step 3. Build dataset objects and scale targets.
A MoleculeDataset wraps the datapoints and applies the chosen featurizer.
Here we used
SimpleMoleculeMolGraphFeaturizer, which turns atoms and bonds into numeric arrays.We also normalized the target values (subtract mean, divide by std) so the model trains smoothly. The stored
scalerallows us to unscale predictions back.
feat = featurizers.SimpleMoleculeMolGraphFeaturizer()
train_set = data.MoleculeDataset(train_dpss[0], featurizer=feat)
scaler = train_set.normalize_targets() # store mean/var
val_set = data.MoleculeDataset(val_dpss[0], featurizer=feat)
val_set.normalize_targets(scaler)
test_set = data.MoleculeDataset(test_dpss[0], featurizer=feat)
# Peek at one item structure
item0 = train_set[0]
type(item0).__name__, item0.y, item0.mg.V.shape, item0.mg.E.shape
('Datum', array([0.42734608]), (11, 72), (24, 14))
#uncomment below and take a look!
#val_set [0]
Step 4. Dataloaders.
Finally, we wrapped datasets in PyTorch-style DataLoaders.
Training loader will shuffle each epoch.
Validation and test loaders do not shuffle, to keep evaluation consistent.
Batching is automatic: molecules of different sizes are packed together and masks are used internally.
train_loader = data.build_dataloader(train_set, num_workers=0)
val_loader = data.build_dataloader(val_set, num_workers=0, shuffle=False)
test_loader = data.build_dataloader(test_set, num_workers=0, shuffle=False)
train_loader
<torch.utils.data.dataloader.DataLoader at 0x797c8a4251f0>
3. Property prediction (regression)#
We will configure a small MPNN for regression.
In particular, we will:
Choose the neural blocks that define how messages are passed, pooled, and transformed into outputs.
Assemble them into a complete model object.
Set up a training loop with early stopping and checkpoints.
Evaluate predictions on a held-out test set and visualize the quality using a parity plot.
3.1 Pick blocks#
mp = nn.BondMessagePassing() # node/edge update
agg = nn.MeanAggregation() # pool node vectors
out = nn.RegressionFFN( # simple FFN head
output_transform=nn.UnscaleTransform.from_standard_scaler(scaler)
)
batch_norm = True
metrics = [nn.metrics.RMSE(), nn.metrics.MAE()] # first metric used for early stopping
BondMessagePassing() updates hidden states on each directed bond by passing information across neighbors.
MeanAggregation() pools hidden vectors to form atom or molecule-level representations. Other options like sum or attention pooling are possible.
RegressionFFN() is a feed-forward head. Here we attach an
UnscaleTransformso predictions can be mapped back to the original solubility scale.Batch normalization improves stability by normalizing hidden states during training.
Metrics let us monitor training. RMSE (root mean squared error) and MAE (mean absolute error) are both useful, but RMSE is often more sensitive to large errors and is used for early stopping.
3.2 Build model and trainer#
Once the blocks are chosen, we wrap them into a full MPNN model. Chemprop uses PyTorch Lightning under the hood, so we also set up a Trainer:
The ModelCheckpoint callback saves the best version of the model during training, based on validation loss.
The trainer can run on CPU or GPU (
accelerator="auto").
We set epoch number at the beginning of this notebook, you can go all the way up and change this number if you feel it takes too long. You can use 15 here for demonstration, but in practice you might extend this depending on dataset size and convergence.
mpnn_sol = models.MPNN(mp, agg, out, batch_norm, metrics)
checkpoint_dir = Path("checkpoints_sol")
checkpoint_dir.mkdir(exist_ok=True)
ckpt = pl.callbacks.ModelCheckpoint(
dirpath=str(checkpoint_dir), filename="best-{epoch}-{val_loss:.3f}",
monitor="val_loss", mode="min", save_last=True
)
trainer = pl.Trainer(
logger=False, enable_checkpointing=True, accelerator="auto",
devices=1, max_epochs=EPOCHS, # you can mannually put = 15 here to make it faster
callbacks=[ckpt]
)
mpnn_sol
INFO:pytorch_lightning.utilities.rank_zero:GPU available: False, used: False
INFO:pytorch_lightning.utilities.rank_zero:TPU available: False, using: 0 TPU cores
INFO:pytorch_lightning.utilities.rank_zero:HPU available: False, using: 0 HPUs
MPNN(
(message_passing): BondMessagePassing(
(W_i): Linear(in_features=86, out_features=300, bias=False)
(W_h): Linear(in_features=300, out_features=300, bias=False)
(W_o): Linear(in_features=372, out_features=300, bias=True)
(dropout): Dropout(p=0.0, inplace=False)
(tau): ReLU()
(V_d_transform): Identity()
(graph_transform): Identity()
)
(agg): MeanAggregation()
(bn): BatchNorm1d(300, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(predictor): RegressionFFN(
(ffn): MLP(
(0): Sequential(
(0): Linear(in_features=300, out_features=300, bias=True)
)
(1): Sequential(
(0): ReLU()
(1): Dropout(p=0.0, inplace=False)
(2): Linear(in_features=300, out_features=1, bias=True)
)
)
(criterion): MSE(task_weights=[[1.0]])
(output_transform): UnscaleTransform()
)
(X_d_transform): Identity()
(metrics): ModuleList(
(0): RMSE(task_weights=[[1.0]])
(1): MAE(task_weights=[[1.0]])
(2): MSE(task_weights=[[1.0]])
)
)
At this stage, we have a complete pipeline: dataset loaders, model blocks, and a trainer that knows when to save progress.
3.3 Train#
Since we implement everything earlier, now training is as simple as calling fit(). The trainer will:
Iterate over the training loader each epoch.
Evaluate on the validation loader.
Save checkpoints when the validation RMSE improves.
During training, you can monitor validation loss to see whether the model is underfitting, overfitting, or converging as expected.
trainer.fit(mpnn_sol, train_loader, val_loader)
INFO:pytorch_lightning.utilities.rank_zero:Loading `train_dataloader` to estimate number of stepping batches.
INFO:
| Name | Type | Params | Mode
---------------------------------------------------------------
0 | message_passing | BondMessagePassing | 227 K | train
1 | agg | MeanAggregation | 0 | train
2 | bn | BatchNorm1d | 600 | train
3 | predictor | RegressionFFN | 90.6 K | train
4 | X_d_transform | Identity | 0 | train
5 | metrics | ModuleList | 0 | train
---------------------------------------------------------------
318 K Trainable params
0 Non-trainable params
318 K Total params
1.276 Total estimated model params size (MB)
25 Modules in train mode
0 Modules in eval mode
INFO:lightning.pytorch.callbacks.model_summary:
| Name | Type | Params | Mode
---------------------------------------------------------------
0 | message_passing | BondMessagePassing | 227 K | train
1 | agg | MeanAggregation | 0 | train
2 | bn | BatchNorm1d | 600 | train
3 | predictor | RegressionFFN | 90.6 K | train
4 | X_d_transform | Identity | 0 | train
5 | metrics | ModuleList | 0 | train
---------------------------------------------------------------
318 K Trainable params
0 Non-trainable params
318 K Total params
1.276 Total estimated model params size (MB)
25 Modules in train mode
0 Modules in eval mode
INFO:pytorch_lightning.utilities.rank_zero:`Trainer.fit` stopped: `max_epochs=50` reached.
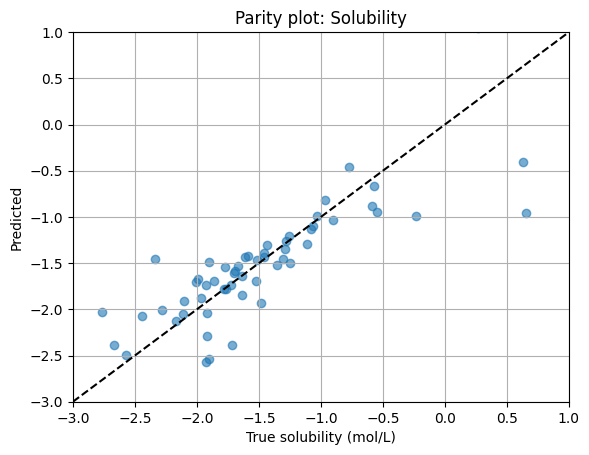
3.4 Test and parity plot#
After training, we hold back the test set for final evaluation. We then visualize predicted vs. true values with a parity plot. We have practiced doing so many times so far.
results = trainer.test(mpnn_sol, test_loader)
# Gather predictions for parity
import torch
with torch.inference_mode():
preds = trainer.predict(mpnn_sol, test_loader)
preds = np.concatenate(preds, axis=0).ravel()
y_true = test_set.Y.ravel()
print("Test size:", len(y_true))
plt.scatter(y_true, preds, alpha=0.6)
# Set both axes to the same range
lims = [-3, 1]
plt.plot(lims, lims, "k--")
plt.xlim(lims)
plt.ylim(lims)
plt.xlabel("True solubility (mol/L)")
plt.ylabel("Predicted")
plt.title("Parity plot: Solubility")
plt.grid(True)
plt.show()
┏━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━━━━━┓ ┃ Test metric ┃ DataLoader 0 ┃ ┡━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━━━━━┩ │ test/mae │ 0.28014591336250305 │ │ test/rmse │ 0.4139021337032318 │ └───────────────────────────┴───────────────────────────┘
Test size: 58
⏰ Exercise
Build a new model call mpnn_sol_2.
Change the aggregation from MeanAggregation() to SumAggregation() and retrain for 10 epochs. Compare RMSE and the parity plot. What changed?
4. Property prediction (classification)#
Here we predict toxic vs non_toxic.
4.1 Build classification dataset#
df_tox = df[["SMILES","tox_bin"]].dropna()
smis = df_tox["SMILES"].tolist()
ys = df_tox["tox_bin"].astype(int).to_numpy().reshape(-1,1)
tox_dps = [data.MoleculeDatapoint.from_smi(s,y) for s,y in zip(smis,ys)]
mols = [dp.mol for dp in tox_dps]
tr_idx, va_idx, te_idx = data.make_split_indices(mols, "random", (0.8,0.1,0.1))
tr, va, te = data.split_data_by_indices(tox_dps, tr_idx, va_idx, te_idx)
feat = featurizers.SimpleMoleculeMolGraphFeaturizer()
tox_tr = data.MoleculeDataset(tr[0], featurizer=feat)
tox_va = data.MoleculeDataset(va[0], featurizer=feat)
tox_te = data.MoleculeDataset(te[0], featurizer=feat)
4.2 Model and training#
mp = nn.BondMessagePassing()
agg = nn.MeanAggregation()
ffn = nn.BinaryClassificationFFN(n_tasks=1)
mpnn_tox = models.MPNN(mp, agg, ffn, batch_norm=False)
tr_loader = data.build_dataloader(tox_tr, num_workers=0)
va_loader = data.build_dataloader(tox_va, num_workers=0, shuffle=False)
te_loader = data.build_dataloader(tox_te, num_workers=0, shuffle=False)
trainer_tox = pl.Trainer(logger=False, enable_checkpointing=True, accelerator="auto",
devices=1, max_epochs=15)
trainer_tox.fit(mpnn_tox, tr_loader, va_loader)
trainer_tox.test(mpnn_tox, te_loader)
INFO:pytorch_lightning.utilities.rank_zero:💡 Tip: For seamless cloud uploads and versioning, try installing [litmodels](https://pypi.org/project/litmodels/) to enable LitModelCheckpoint, which syncs automatically with the Lightning model registry.
INFO:pytorch_lightning.utilities.rank_zero:GPU available: False, used: False
INFO:pytorch_lightning.utilities.rank_zero:TPU available: False, using: 0 TPU cores
INFO:pytorch_lightning.utilities.rank_zero:HPU available: False, using: 0 HPUs
INFO:pytorch_lightning.utilities.rank_zero:Loading `train_dataloader` to estimate number of stepping batches.
INFO:
| Name | Type | Params | Mode
--------------------------------------------------------------------
0 | message_passing | BondMessagePassing | 227 K | train
1 | agg | MeanAggregation | 0 | train
2 | bn | Identity | 0 | train
3 | predictor | BinaryClassificationFFN | 90.6 K | train
4 | X_d_transform | Identity | 0 | train
5 | metrics | ModuleList | 0 | train
--------------------------------------------------------------------
318 K Trainable params
0 Non-trainable params
318 K Total params
1.273 Total estimated model params size (MB)
24 Modules in train mode
0 Modules in eval mode
INFO:lightning.pytorch.callbacks.model_summary:
| Name | Type | Params | Mode
--------------------------------------------------------------------
0 | message_passing | BondMessagePassing | 227 K | train
1 | agg | MeanAggregation | 0 | train
2 | bn | Identity | 0 | train
3 | predictor | BinaryClassificationFFN | 90.6 K | train
4 | X_d_transform | Identity | 0 | train
5 | metrics | ModuleList | 0 | train
--------------------------------------------------------------------
318 K Trainable params
0 Non-trainable params
318 K Total params
1.273 Total estimated model params size (MB)
24 Modules in train mode
0 Modules in eval mode
INFO:pytorch_lightning.utilities.rank_zero:`Trainer.fit` stopped: `max_epochs=15` reached.
┏━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━━━━━┓ ┃ Test metric ┃ DataLoader 0 ┃ ┡━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━━━━━┩ │ test/roc │ 0.8639456033706665 │ └───────────────────────────┴───────────────────────────┘
[{'test/roc': 0.8639456033706665}]
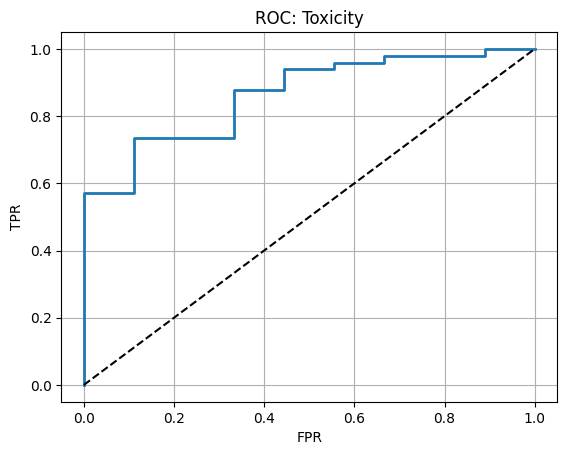
4.3 ROC curve#
# Gather probabilities
with torch.inference_mode():
pred_chunks = trainer_tox.predict(mpnn_tox, te_loader)
proba = np.concatenate(pred_chunks, axis=0).ravel()
y_true = tox_te.Y.ravel().astype(int)
auc = roc_auc_score(y_true, proba)
acc = accuracy_score(y_true, (proba>=0.5).astype(int))
print(f"Test AUC: {auc:.3f} Accuracy: {acc:.3f}")
fpr, tpr, thr = roc_curve(y_true, proba)
plt.plot(fpr, tpr, lw=2)
plt.plot([0,1],[0,1],"k--")
plt.xlabel("FPR")
plt.ylabel("TPR")
plt.title("ROC: Toxicity")
plt.grid(True)
plt.show()
Test AUC: 0.864 Accuracy: 0.862
5. Reactivity and selectivity#
Two targets in this dataset relate to reactions.
Reactivity: binary at the molecule level.
Selectivity: oxidation site indices at the atom level.
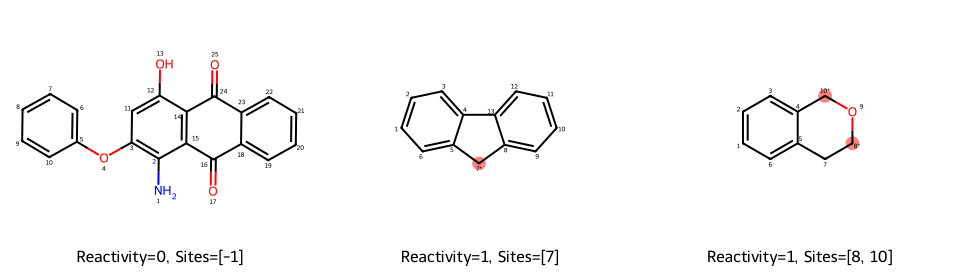
Before we train models, let’s look at the labels. Let’s pick three representative molecules from the C-H oxidation dataset and see their reactivity and selectivity label.
# Helper selection: one negative (no reaction), one positive with a single site, one positive with multiple sites
def pick_representatives(df):
df_pos = df[(df["react_bin"] == 1) & (df["site_list"].map(len) >= 1)].copy()
df_pos_multi = df_pos[df_pos["site_list"].map(len) >= 2].copy()
df_neg = df[(df["react_bin"] == 0)].copy()
reps = []
if not df_neg.empty:
reps.append(("Negative (react_bin=0)", df_neg.iloc[0]))
if not df_pos.empty:
reps.append(("Positive (react_bin=1; 1 site)", df_pos[df_pos["site_list"].map(len) == 1].iloc[0]
if (df_pos["site_list"].map(len) == 1).any() else df_pos.iloc[0]))
if not df_pos_multi.empty:
reps.append(("Positive (react_bin=1; multi-site)", df_pos_multi.iloc[0]))
# If fewer than 3 examples exist, just return what we have
return reps
reps = pick_representatives(df)
len(reps), [t for t,_ in reps]
# Show the chosen rows so readers see SMILES and labels
import pandas as pd
def row_view(r):
return {
"SMILES": r["SMILES"],
"react_bin": r["react_bin"],
"site_list (1-based)": r["site_list"]
}
rep_table = pd.DataFrame([row_view(row) for _, row in reps])
rep_table
| SMILES | react_bin | site_list (1-based) | |
|---|---|---|---|
| 0 | Nc1c(Oc2ccccc2)cc(O)c2c1C(=O)c1ccccc1C2=O | 0 | [-1] |
| 1 | c1ccc2c(c1)Cc1ccccc1-2 | 1 | [7] |
| 2 | c1ccc2c(c1)CCOC2 | 1 | [8, 10] |
Let’s draw them:
from rdkit.Chem.Draw import rdMolDraw2D
def make_annotated_copy(mol, site_list_1based=None, tag_c123=True):
m = Chem.Mol(mol) # copy
Chem.AssignAtomChiralTagsFromStructure(m)
Chem.Kekulize(m, clearAromaticFlags=True)
n = m.GetNumAtoms()
# Highlight oxidation sites (convert to 0-based safely)
hi_atoms = []
if site_list_1based:
for idx1 in site_list_1based:
j = idx1 - 1
if 0 <= j < n:
hi_atoms.append(j)
# Always annotate the atom index so readers see 1-based indexing used in labels
for j in range(n):
a = m.GetAtomWithIdx(j)
idx1 = j + 1
old = a.GetProp("atomNote") if a.HasProp("atomNote") else ""
# If this atom is an oxidation site, add a star
star = "*" if (j in hi_atoms) else ""
a.SetProp("atomNote", f"{old} {idx1}{star}".strip())
return m, hi_atoms
def draw_examples(reps, mol_size=(320, 280)):
ms = []
legends = []
highlights = []
for title, row in reps:
smi = row["SMILES"]
m = Chem.MolFromSmiles(smi)
if m is None:
continue
m_annot, hi = make_annotated_copy(m, site_list_1based=row["site_list"], tag_c123=True)
ms.append(m_annot)
lbl = f"Reactivity={row['react_bin']}, Sites={row['site_list']}"
legends.append(lbl)
highlights.append(hi)
imgs = []
for m, hi, lg in zip(ms, highlights, legends):
img = Draw.MolToImage(m, size=mol_size, highlightAtoms=hi)
img.info["legend"] = lg
imgs.append(img)
# Create a grid image manually by re-drawing with legends
return Draw.MolsToGridImage(ms, molsPerRow=len(ms), subImgSize=mol_size,
legends=legends,
highlightAtomLists=highlights)
grid_img = draw_examples(reps)
grid_img
5.1 Reactivity classifier#
This mirrors the toxicity classification workflow we saw before.
df_rxn = df[["SMILES","react_bin"]].dropna()
smis = df_rxn["SMILES"].tolist()
ys = df_rxn["react_bin"].astype(int).to_numpy().reshape(-1,1)
rxn_dps = [data.MoleculeDatapoint.from_smi(s,y) for s,y in zip(smis, ys)]
mols = [dp.mol for dp in rxn_dps]
tr_idx, va_idx, te_idx = data.make_split_indices(mols, "random", (0.8,0.1,0.1))
tr, va, te = data.split_data_by_indices(rxn_dps, tr_idx, va_idx, te_idx)
feat = featurizers.SimpleMoleculeMolGraphFeaturizer()
rxn_tr = data.MoleculeDataset(tr[0], featurizer=feat)
rxn_va = data.MoleculeDataset(va[0], featurizer=feat)
rxn_te = data.MoleculeDataset(te[0], featurizer=feat)
mp = nn.BondMessagePassing()
agg = nn.MeanAggregation()
ffn = nn.BinaryClassificationFFN(n_tasks=1)
mpnn_rxn = models.MPNN(mp, agg, ffn, batch_norm=False)
tr_loader = data.build_dataloader(rxn_tr, num_workers=0)
va_loader = data.build_dataloader(rxn_va, num_workers=0, shuffle=False)
te_loader = data.build_dataloader(rxn_te, num_workers=0, shuffle=False)
trainer_rxn = pl.Trainer(logger=False, enable_checkpointing=True, accelerator="auto",
devices=1, max_epochs=15)
trainer_rxn.fit(mpnn_rxn, tr_loader, va_loader)
trainer_rxn.test(mpnn_rxn, te_loader)
INFO:pytorch_lightning.utilities.rank_zero:💡 Tip: For seamless cloud uploads and versioning, try installing [litmodels](https://pypi.org/project/litmodels/) to enable LitModelCheckpoint, which syncs automatically with the Lightning model registry.
INFO:pytorch_lightning.utilities.rank_zero:GPU available: False, used: False
INFO:pytorch_lightning.utilities.rank_zero:TPU available: False, using: 0 TPU cores
INFO:pytorch_lightning.utilities.rank_zero:HPU available: False, using: 0 HPUs
INFO:pytorch_lightning.utilities.rank_zero:Loading `train_dataloader` to estimate number of stepping batches.
INFO:
| Name | Type | Params | Mode
--------------------------------------------------------------------
0 | message_passing | BondMessagePassing | 227 K | train
1 | agg | MeanAggregation | 0 | train
2 | bn | Identity | 0 | train
3 | predictor | BinaryClassificationFFN | 90.6 K | train
4 | X_d_transform | Identity | 0 | train
5 | metrics | ModuleList | 0 | train
--------------------------------------------------------------------
318 K Trainable params
0 Non-trainable params
318 K Total params
1.273 Total estimated model params size (MB)
24 Modules in train mode
0 Modules in eval mode
INFO:lightning.pytorch.callbacks.model_summary:
| Name | Type | Params | Mode
--------------------------------------------------------------------
0 | message_passing | BondMessagePassing | 227 K | train
1 | agg | MeanAggregation | 0 | train
2 | bn | Identity | 0 | train
3 | predictor | BinaryClassificationFFN | 90.6 K | train
4 | X_d_transform | Identity | 0 | train
5 | metrics | ModuleList | 0 | train
--------------------------------------------------------------------
318 K Trainable params
0 Non-trainable params
318 K Total params
1.273 Total estimated model params size (MB)
24 Modules in train mode
0 Modules in eval mode
INFO:pytorch_lightning.utilities.rank_zero:`Trainer.fit` stopped: `max_epochs=15` reached.
┏━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━━━━━┓ ┃ Test metric ┃ DataLoader 0 ┃ ┡━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━━━━━┩ │ test/roc │ 0.958184003829956 │ └───────────────────────────┴───────────────────────────┘
[{'test/roc': 0.958184003829956}]
import ast
def atoms_labels_from_smiles(smi, positive_idxs):
if Chem is None:
return None
m = Chem.MolFromSmiles(smi)
if m is None:
return None
n = m.GetNumAtoms()
y = np.zeros((n,1), dtype=float)
for idx in positive_idxs:
# dataset uses 1-based indexing in the text, RDKit uses 0-based
j = idx-1
if 0 <= j < n:
y[j,0] = 1.0
return y
# Build list of MolAtomBondDatapoint for selectivity
sel_rows = df[["SMILES","site_list"]].dropna()
sel_dps = []
for smi, sites in sel_rows.itertuples(index=False):
atom_y = atoms_labels_from_smiles(smi, sites)
if atom_y is None:
continue
# We provide atom_y, molecule-level y is optional here
dp = data.MolAtomBondDatapoint.from_smi(
smi, atom_y=atom_y, reorder_atoms=False
)
sel_dps.append(dp)
len(sel_dps), type(sel_dps[0]).__name__
(575, 'MolAtomBondDatapoint')
mols = [Chem.MolFromSmiles(dp.name) if hasattr(dp, "name") else None for dp in sel_dps]
# For structure-based split we need RDKit Mol. Build directly from SMILES fallback:
mols = [Chem.MolFromSmiles(df.loc[df["SMILES"]==dp.name, "SMILES"].iloc[0]) if Chem else None for dp in sel_dps]
tr_idx, va_idx, te_idx = data.make_split_indices(mols, "random", (0.8, 0.1, 0.1))
tr, va, te = data.split_data_by_indices(sel_dps, tr_idx, va_idx, te_idx)
feat = featurizers.SimpleMoleculeMolGraphFeaturizer()
tr_set = data.MolAtomBondDataset(tr[0], featurizer=feat)
va_set = data.MolAtomBondDataset(va[0], featurizer=feat)
te_set = data.MolAtomBondDataset(te[0], featurizer=feat)
tr_loader = data.build_dataloader(tr_set, shuffle=True, batch_size=8)
va_loader = data.build_dataloader(va_set, shuffle=False, batch_size=8)
te_loader = data.build_dataloader(te_set, shuffle=False, batch_size=8)
WARNING:chemprop.data.dataloader:Dropping last batch of size 1 to avoid issues with batch normalization (dataset size = 57, batch_size = 8)
mp = nn.MABBondMessagePassing(
d_v=feat.atom_fdim, d_e=feat.bond_fdim, d_h=300, depth=3, dropout=0.1
)
agg = nn.MeanAggregation()
atom_predictor = nn.BinaryClassificationFFN(n_tasks=1) # atom-level 0/1
model_sel = models.MolAtomBondMPNN(
message_passing=mp,
agg=agg,
mol_predictor=None,
atom_predictor=atom_predictor,
bond_predictor=None,
batch_norm=True,
metrics=[nn.metrics.BinaryAUROC()],
)
trainer_sel = pl.Trainer(logger=False, enable_checkpointing=True, accelerator="auto",
devices=1, max_epochs= EPOCHS) #You can change this number at the begining of this notebook
trainer_sel.fit(model_sel, tr_loader, va_loader)
trainer_sel.test(model_sel, te_loader)
INFO:pytorch_lightning.utilities.rank_zero:💡 Tip: For seamless cloud uploads and versioning, try installing [litmodels](https://pypi.org/project/litmodels/) to enable LitModelCheckpoint, which syncs automatically with the Lightning model registry.
INFO:pytorch_lightning.utilities.rank_zero:GPU available: False, used: False
INFO:pytorch_lightning.utilities.rank_zero:TPU available: False, using: 0 TPU cores
INFO:pytorch_lightning.utilities.rank_zero:HPU available: False, using: 0 HPUs
INFO:pytorch_lightning.utilities.rank_zero:Loading `train_dataloader` to estimate number of stepping batches.
INFO:
| Name | Type | Params | Mode
--------------------------------------------------------------------
0 | message_passing | MABBondMessagePassing | 322 K | train
1 | agg | MeanAggregation | 0 | train
2 | atom_predictor | BinaryClassificationFFN | 90.6 K | train
3 | bns | ModuleList | 600 | train
4 | X_d_transform | Identity | 0 | train
5 | metricss | ModuleList | 0 | train
--------------------------------------------------------------------
413 K Trainable params
0 Non-trainable params
413 K Total params
1.654 Total estimated model params size (MB)
30 Modules in train mode
0 Modules in eval mode
INFO:lightning.pytorch.callbacks.model_summary:
| Name | Type | Params | Mode
--------------------------------------------------------------------
0 | message_passing | MABBondMessagePassing | 322 K | train
1 | agg | MeanAggregation | 0 | train
2 | atom_predictor | BinaryClassificationFFN | 90.6 K | train
3 | bns | ModuleList | 600 | train
4 | X_d_transform | Identity | 0 | train
5 | metricss | ModuleList | 0 | train
--------------------------------------------------------------------
413 K Trainable params
0 Non-trainable params
413 K Total params
1.654 Total estimated model params size (MB)
30 Modules in train mode
0 Modules in eval mode
INFO:pytorch_lightning.utilities.rank_zero:`Trainer.fit` stopped: `max_epochs=50` reached.
┏━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━━━━━┓ ┃ Test metric ┃ DataLoader 0 ┃ ┡━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━━━━━┩ │ atom_test/roc │ 0.8670018315315247 │ └───────────────────────────┴───────────────────────────┘
[{'atom_test/roc': 0.8670018315315247}]
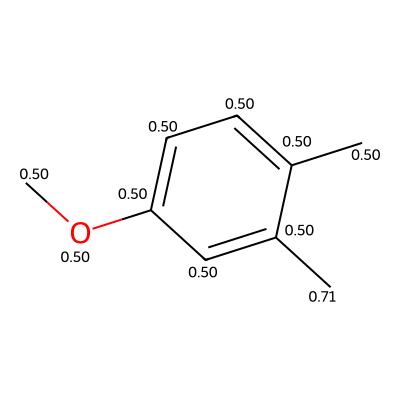
# 1) Pick which test item to visualize
idx = 11 # change as you like
# 2) Grab the original datapoint (not the already-featurized datum)
dp = te_set.data[idx] # IMPORTANT: .data gives you the raw MolAtomBondDatapoint
# 3) Make a one-item MolAtomBondDataset with the SAME featurizer you used before
single_ds = data.MolAtomBondDataset([dp], featurizer=feat)
# 4) Build a loader and get its batch
single_loader = data.build_dataloader(single_ds, shuffle=False, batch_size=1)
batch = next(iter(single_loader)) # this now matches collate expectations
with torch.inference_mode():
out = model_sel(batch[0], None) # batch[0] is the MolGraphBatch
# 6) Unpack outputs and get per-atom probabilities
_, atom_logits, _ = out
atom_probs = torch.sigmoid(atom_logits).cpu().numpy().ravel()
print("Atom count:", len(atom_probs))
print("First 10 probabilities:", atom_probs[:10])
# 7) Draw the SAME molecule with aligned probabilities
smi = dp.name # MolAtomBondDatapoint stores SMILES in .name
m = Chem.MolFromSmiles(smi)
m2 = Chem.Mol(m)
for i, a in enumerate(m2.GetAtoms()):
a.SetProp("atomNote", f"{atom_probs[i]:.2f}")
img = Draw.MolToImage(m2, size=(400, 400))
display(img)
Atom count: 10
First 10 probabilities: [0.5 0.5000001 0.5 0.5 0.5000006 0.5000006
0.503788 0.5000002 0.7055641 0.50000006]
⏰ Exercise
Change idx above to take a look at other molecules in test set.
6. Rationale#
In this section we ask why the model makes its predictions. Instead of just reporting a number for solubility or reactivity, we try to find the substructures of a molecule that drive the prediction.
The method we use is a simplified form of Monte Carlo Tree Search (MCTS) rationale extraction from the Chemprop:
Start with the whole molecule and its predicted property.
Systematically remove small fragments (like a bond or a ring) and check how the model’s prediction changes for the remaining subgraph.
Build a search tree of candidate subgraphs and keep the ones that still predict strongly for the property of interest.
From this set, report the smallest and most predictive subgraph as the rationale.
# ===== Shared helpers for both tasks (Chemprop v2.2.1) =====
from dataclasses import dataclass, field
from typing import Callable, Iterable, List, Tuple, Union
import math, time
import numpy as np
import pandas as pd
import torch
from lightning import pytorch as pl
from rdkit import Chem
from rdkit.Chem.Draw import rdMolDraw2D
from IPython.display import display, Image
from chemprop import data, models
from chemprop.models import MPNN
# Predict a list of SMILES with a single-task model. Returns array shape (n, 1).
def make_prediction(models_in: List[MPNN], trainer_in: pl.Trainer, smiles: List[str]) -> np.ndarray:
test_data = [data.MoleculeDatapoint.from_smi(s) for s in smiles]
test_dset = data.MoleculeDataset(test_data)
test_loader = data.build_dataloader(test_dset, batch_size=1, num_workers=0, shuffle=False)
with torch.inference_mode():
agg = None
for m in models_in:
pred_batches = trainer_in.predict(m, test_loader) # list of tensors/arrays
preds = torch.cat([torch.as_tensor(pb) for pb in pred_batches], dim=0).cpu().numpy()
agg = preds if agg is None else (agg + preds)
return agg / len(models_in)
# ---- MCTS components (as in tutorial, with safer kekulize handling) ----
@dataclass
class MCTSNode:
smiles: str
atoms: Iterable[int]
W: float = 0
N: float = 0
P: float = 0
children: list = field(default_factory=list)
def __post_init__(self): self.atoms = set(self.atoms)
def Q(self) -> float: return self.W / self.N if self.N > 0 else 0
def U(self, n: int, c_puct: float = 10.0) -> float: return c_puct * self.P * math.sqrt(n) / (1 + self.N)
def find_clusters(mol: Chem.Mol) -> Tuple[List[Tuple[int,...]], List[List[int]]]:
n = mol.GetNumAtoms()
if n == 1: return [(0,)], [[0]]
clusters = []
for b in mol.GetBonds():
i, j = b.GetBeginAtomIdx(), b.GetEndAtomIdx()
if not b.IsInRing():
clusters.append((i, j))
ssr = [tuple(x) for x in Chem.GetSymmSSSR(mol)]
clusters.extend(ssr)
atom_cls = [[] for _ in range(n)]
for k, cl in enumerate(clusters):
for a in cl: atom_cls[a].append(k)
return clusters, atom_cls
def extract_subgraph_from_mol(mol: Chem.Mol, selected_atoms: set) -> Tuple[Chem.Mol, List[int]]:
sel = set(selected_atoms)
roots = []
for idx in sel:
atom = mol.GetAtomWithIdx(idx)
if any(nei.GetIdx() not in sel for nei in atom.GetNeighbors()):
roots.append(idx)
rw = Chem.RWMol(mol)
for idx in roots:
a = rw.GetAtomWithIdx(idx)
a.SetAtomMapNum(1)
aroma = [b for b in a.GetBonds() if b.GetBondType() == Chem.rdchem.BondType.AROMATIC]
aroma = [b for b in aroma if b.GetBeginAtom().GetIdx() in sel and b.GetEndAtom().GetIdx() in sel]
if len(aroma) == 0: a.SetIsAromatic(False)
for idx in sorted([a.GetIdx() for a in rw.GetAtoms() if a.GetIdx() not in sel], reverse=True):
rw.RemoveAtom(idx)
return rw.GetMol(), roots
def extract_subgraph(smiles: str, selected_atoms: set) -> Tuple[Union[str,None], Union[List[int],None]]:
mol = Chem.MolFromSmiles(smiles)
if mol is None: return None, None
# try kekulized
try:
mk = Chem.Mol(mol); Chem.Kekulize(mk)
sub, roots = extract_subgraph_from_mol(mk, selected_atoms)
try:
smi = Chem.MolToSmiles(sub, kekuleSmiles=True)
sub = Chem.MolFromSmiles(smi)
except Exception: sub = None
if sub is not None and mol.HasSubstructMatch(sub): return Chem.MolToSmiles(sub), roots
except Exception:
pass
# fallback without kekulize
sub, roots = extract_subgraph_from_mol(mol, selected_atoms)
try:
smi = Chem.MolToSmiles(sub)
sub = Chem.MolFromSmiles(smi)
except Exception: sub = None
return (Chem.MolToSmiles(sub), roots) if sub is not None else (None, None)
def mcts_rollout(node: MCTSNode, state_map, orig_smiles, clusters, atom_cls, nei_cls,
scoring_fn: Callable[[List[str]], np.ndarray], min_atoms=8, c_puct=10.0) -> float:
cur = node.atoms
if len(cur) <= min_atoms: return node.P
if len(node.children) == 0:
cur_cls = set([i for i, x in enumerate(clusters) if x <= cur])
for i in cur_cls:
leaf_atoms = [a for a in clusters[i] if len(atom_cls[a] & cur_cls) == 1]
if len(nei_cls[i] & cur_cls) == 1 or (len(clusters[i]) == 2 and len(leaf_atoms) == 1):
new_atoms = cur - set(leaf_atoms)
new_smi, _ = extract_subgraph(orig_smiles, new_atoms)
if not new_smi: continue
node.children.append(state_map.get(new_smi, MCTSNode(new_smi, new_atoms)))
state_map[node.smiles] = node
if len(node.children) == 0: return node.P
scores = scoring_fn([x.smiles for x in node.children])
for child, sc in zip(node.children, scores): child.P = float(sc)
totalN = sum(c.N for c in node.children)
nxt = max(node.children, key=lambda x: x.Q() + x.U(totalN, c_puct=c_puct))
v = mcts_rollout(nxt, state_map, orig_smiles, clusters, atom_cls, nei_cls, scoring_fn, min_atoms=min_atoms, c_puct=c_puct)
nxt.W += v; nxt.N += 1
return v
def run_mcts_for_smiles(smi: str, scoring_fn, rollout=10, c_puct=10.0,
min_atoms=8, max_atoms=20) -> List[MCTSNode]:
mol = Chem.MolFromSmiles(smi)
if mol is None: return []
clusters_raw, atom_cls_raw = find_clusters(mol)
clusters = [set(cl) for cl in clusters_raw]
atom_cls = [set(x) for x in atom_cls_raw]
nei_cls = []
for i, cl in enumerate(clusters):
neigh = [nei for a in cl for nei in atom_cls_raw[a]]
nei_cls.append(set(neigh) - {i})
root = MCTSNode(smi, set(range(mol.GetNumAtoms())))
state_map = {smi: root}
for _ in range(rollout):
mcts_rollout(root, state_map, smi, clusters, atom_cls, nei_cls, scoring_fn, min_atoms=min_atoms, c_puct=c_puct)
rats = [node for _, node in state_map.items() if node.smiles is not None and len(node.atoms) <= max_atoms]
return rats
# Simple fragment fallback if MCTS cannot find anything
def find_fragments(mol: Chem.Mol):
frags = []
for b in mol.GetBonds():
if not b.IsInRing():
frags.append({b.GetBeginAtomIdx(), b.GetEndAtomIdx()})
try:
for ring in Chem.GetSymmSSSR(mol):
frags.append(set(int(i) for i in ring))
except Exception:
pass
return frags
def fragment_top1(smi: str, scoring_fn) -> List[Tuple[str, float]]:
mol = Chem.MolFromSmiles(smi)
if mol is None: return []
cands = []
for aset in find_fragments(mol):
sub_smi, _ = extract_subgraph(smi, aset)
if sub_smi: cands.append(sub_smi)
if not cands: return []
cands = list(dict.fromkeys(cands))
scores = scoring_fn(cands)
idx = int(np.argmax(scores))
return [(cands[idx], float(scores[idx]))]
def visualize_rationale_on_parent(parent_smi: str, rationale_smi: str, size=(520, 420)):
pm = Chem.MolFromSmiles(parent_smi)
rm = Chem.MolFromSmiles(rationale_smi)
if pm is None or rm is None:
print("Cannot draw: invalid SMILES.")
return
match = pm.GetSubstructMatch(rm)
hi_atoms = list(match) if match else []
drawer = rdMolDraw2D.MolDraw2DCairo(size[0], size[1])
opts = drawer.drawOptions()
for attr in ("atomLabelFontSize", "fontSize", "scalingFactor"):
if hasattr(opts, attr):
try:
setattr(opts, attr, 0.9)
break
except Exception:
pass
rdMolDraw2D.PrepareAndDrawMolecule(drawer, pm,
highlightAtoms=hi_atoms if hi_atoms else None)
drawer.FinishDrawing()
png = drawer.GetDrawingText()
display(Image(data=png))
Because scoring is done by predicting each subgraph as if it were a standalone molecule, the absolute values (like negative logS) may not be directly comparable to the parent’s score. What matters is the difference (Δ): how much the rationale’s score differs from the parent. A positive Δ means the subgraph supports the property; a negative Δ means removing that part lowers the property.
We generate a summary table with one rationale (rationale_0) for each molecule:
smiles: the original moleculelogSorreact_prob: the model’s prediction on the full moleculerationale_0: the SMILES string of the key substructurerationale_0_score: the model’s prediction on that substructurerationale_0_delta: the difference between rationale and parent
Finally, we visualize the rationale by highlighting the matching atoms in the parent molecule. This lets us
6.1 Solubility prediction rationale#
# ===== Solubility summary table with top-1 rationale =====
# Uses your trained mpnn_sol and trainer, and df_sol["SMILES"]
models_sol = [mpnn_sol] # must be single-task regression
prop_name = "logS"
def scoring_fn_sol(smiles_list: List[str]) -> np.ndarray:
return make_prediction(models_sol, trainer, smiles_list)[:, 0]
# Build table
N = 15 # how many molecules to summarize
sample_smis = df_sol["SMILES"].tolist()[:N]
rows = {"smiles": [], prop_name: [], "rationale_0": [], "rationale_0_score": []}
t0 = time.time()
for smi in sample_smis:
base = float(scoring_fn_sol([smi])[0])
rows["smiles"].append(smi)
rows[prop_name].append(base)
# size-aware MCTS settings
mol = Chem.MolFromSmiles(smi)
n = mol.GetNumAtoms() if mol is not None else 20
min_atoms = max(3, int(0.30 * n))
max_atoms = max(min_atoms + 1, int(0.60 * n))
rats = run_mcts_for_smiles(smi, scoring_fn_sol, rollout=10, c_puct=10.0,
min_atoms=min_atoms, max_atoms=max_atoms)
if rats:
# choose smallest subgraphs, then highest score
ms = min(len(x.atoms) for x in rats)
kept = [x for x in rats if len(x.atoms) == ms and x.smiles is not None]
kept.sort(key=lambda x: x.P, reverse=True)
r0 = kept[0].smiles
r0s = float(kept[0].P)
else:
# fallback to fragment top1
top1 = fragment_top1(smi, scoring_fn_sol)
if top1:
r0, r0s = top1[0]
else:
r0, r0s = None, None
rows["rationale_0"].append(r0)
rows["rationale_0_score"].append(r0s)
elapsed = time.time() - t0
print(f"Solubility: built table for {len(sample_smis)} molecules in {elapsed:.2f}s")
solubility_rationales = pd.DataFrame(rows)
solubility_rationales
Solubility: built table for 15 molecules in 3.48s
| smiles | logS | rationale_0 | rationale_0_score | |
|---|---|---|---|---|
| 0 | c1ccc2c(c1)CCOC2 | -1.195756 | C1C[CH:1]=[CH:1]CO1 | -0.505139 |
| 1 | c1ccc2c(c1)Cc1ccccc1-2 | -1.719822 | c1cc[cH:1][cH:1]c1 | -0.771889 |
| 2 | c1ccc2c(c1)CCCC2 | -1.534268 | C1CC[CH:1]=[CH:1]C1 | -1.092782 |
| 3 | CCc1ccccc1 | -1.301932 | C[CH3:1] | -0.297537 |
| 4 | C1=CCCCC1 | -1.092782 | C1=CCCCC1 | -1.092782 |
| 5 | C1CCSC1 | -0.878732 | C1CCSC1 | -0.878732 |
| 6 | CN1CCCC1=O | -0.318836 | O=[CH2:1] | 1.264910 |
| 7 | COCc1ccccc1 | -1.092736 | COC[CH3:1] | -0.250766 |
| 8 | CCCCN1Cc2ccccc2C1 | -1.399117 | C1[CH:1]=[CH:1]C[NH:1]1 | 0.487650 |
| 9 | CCCc1ccccc1 | -1.500662 | C[CH3:1] | -0.297537 |
| 10 | CCCCCCCCOCc1ccc(OC)cc1 | -2.007931 | c1c[cH:1]cc[cH:1]1 | -0.771889 |
| 11 | CC(C)CCOCc1ccccc1 | -1.739935 | C(O[CH3:1])[CH3:1] | -0.250766 |
| 12 | CC1=CCCCC1 | -1.427984 | C[CH3:1] | -0.297537 |
| 13 | CC1=C(C)CCCC1 | -1.500649 | C[CH3:1] | -0.297537 |
| 14 | C1=CCCC1 | -1.037244 | C1=CCCC1 | -1.037244 |
if pd.notna(solubility_rationales.loc[0, "rationale_0"]):
parent = solubility_rationales.loc[0, "smiles"]
rat = solubility_rationales.loc[0, "rationale_0"]
print("Parent:", parent)
print("Rationale 0:", rat, " score=", solubility_rationales.loc[0, "rationale_0_score"])
visualize_rationale_on_parent(parent, rat)
Parent: c1ccc2c(c1)CCOC2
Rationale 0: C1C[CH:1]=[CH:1]CO1 score= -0.5051385164260864
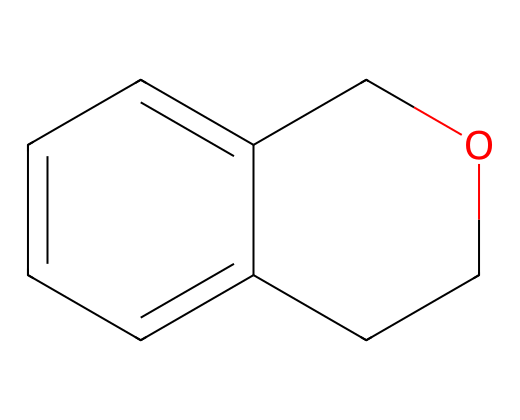
6.2 Reactivity prediction rationale#
# ===== Reactivity summary table with top-1 rationale =====
# Uses your trained mpnn_rxn and trainer_rxn, and df_rxn["SMILES"]
models_rxn = [mpnn_rxn] # single-task binary classification head
trainer_cls = trainer_rxn # your classification trainer
def scoring_fn_rxn(smiles_list: List[str]) -> np.ndarray:
# predict logits, convert to probabilities with sigmoid
preds = make_prediction(models_rxn, trainer_cls, smiles_list)[:, 0]
# Some builds return probs already; to be safe, apply sigmoid if outside [0,1]
if ((preds < 0) | (preds > 1)).any():
preds = 1.0 / (1.0 + np.exp(-preds))
return preds.reshape(-1,)
# Build table
N = 15
sample_smis_rxn = df_rxn["SMILES"].tolist()[:N]
rows_rxn = {"smiles": [], "react_prob": [], "rationale_0": [], "rationale_0_score": []}
t0 = time.time()
for smi in sample_smis_rxn:
base_p = float(scoring_fn_rxn([smi])[0])
rows_rxn["smiles"].append(smi)
rows_rxn["react_prob"].append(base_p)
mol = Chem.MolFromSmiles(smi)
n = mol.GetNumAtoms() if mol is not None else 20
min_atoms = max(3, int(0.30 * n))
max_atoms = max(min_atoms + 1, int(0.60 * n))
rats = run_mcts_for_smiles(smi, scoring_fn_rxn, rollout=10, c_puct=10.0,
min_atoms=min_atoms, max_atoms=max_atoms)
if rats:
ms = min(len(x.atoms) for x in rats)
kept = [x for x in rats if len(x.atoms) == ms and x.smiles is not None]
kept.sort(key=lambda x: x.P, reverse=True)
r0 = kept[0].smiles
r0s = float(kept[0].P)
else:
top1 = fragment_top1(smi, scoring_fn_rxn)
if top1:
r0, r0s = top1[0]
else:
r0, r0s = None, None
rows_rxn["rationale_0"].append(r0)
rows_rxn["rationale_0_score"].append(r0s)
elapsed = time.time() - t0
print(f"Reactivity: built table for {len(sample_smis_rxn)} molecules in {elapsed:.2f}s")
reactivity_rationales = pd.DataFrame(rows_rxn)
reactivity_rationales
Reactivity: built table for 15 molecules in 6.96s
| smiles | react_prob | rationale_0 | rationale_0_score | |
|---|---|---|---|---|
| 0 | c1ccc2c(c1)CCOC2 | 0.944441 | C1C[CH:1]=[CH:1]CO1 | 0.916721 |
| 1 | c1ccc2c(c1)Cc1ccccc1-2 | 0.867228 | C1[CH:1]=[CH:1][CH:1]=[CH:1]1 | 0.841032 |
| 2 | c1ccc2c(c1)CCCC2 | 0.966502 | C1CC[CH:1]=[CH:1]C1 | 0.960123 |
| 3 | CCc1ccccc1 | 0.908582 | c1cc[cH:1]cc1 | 0.825776 |
| 4 | C1=CCCCC1 | 0.960123 | C1=CCCCC1 | 0.960123 |
| 5 | C1CCSC1 | 0.963128 | C1CCSC1 | 0.963128 |
| 6 | CN1CCCC1=O | 0.877834 | C1C[CH2:1][NH:1]C1 | 0.958719 |
| 7 | COCc1ccccc1 | 0.876020 | COC[CH3:1] | 0.841456 |
| 8 | CCCCN1Cc2ccccc2C1 | 0.945808 | CCC[CH3:1] | 0.916729 |
| 9 | CCCc1ccccc1 | 0.923266 | CCC[CH3:1] | 0.916729 |
| 10 | CCCCCCCCOCc1ccc(OC)cc1 | 0.920641 | C(CC[CH3:1])C[CH3:1] | 0.949148 |
| 11 | CC(C)CCOCc1ccccc1 | 0.906913 | C[CH2:1]C[CH3:1] | 0.916729 |
| 12 | CC1=CCCCC1 | 0.958145 | C1=[CH:1]CCCC1 | 0.960123 |
| 13 | CC1=C(C)CCCC1 | 0.955647 | C1CC[CH:1]=[CH:1]C1 | 0.960123 |
| 14 | C1=CCCC1 | 0.952870 | C1=CCCC1 | 0.952870 |
if pd.notna(reactivity_rationales.loc[0, "rationale_0"]):
parent = reactivity_rationales.loc[0, "smiles"]
rat = reactivity_rationales.loc[0, "rationale_0"]
print("Parent:", parent)
print("Rationale 0:", rat, " score=", reactivity_rationales.loc[0, "rationale_0_score"])
visualize_rationale_on_parent(parent, rat)
Parent: c1ccc2c(c1)CCOC2
Rationale 0: C1C[CH:1]=[CH:1]CO1 score= 0.9167211055755615

7. Chemprop CLI (Command-Line Interface)#
Prepare a minimal CSV: SMILES,Melting Point.
# Load data and write a small CSV
url = "https://raw.githubusercontent.com/zzhenglab/ai4chem/main/book/_data/C_H_oxidation_dataset.csv"
df = pd.read_csv(url)
reg_cols = ["SMILES", "Melting Point"]
df_reg = df[reg_cols].dropna().copy()
df_reg.head(3)
| SMILES | Melting Point | |
|---|---|---|
| 0 | c1ccc2c(c1)CCOC2 | 65.8 |
| 1 | c1ccc2c(c1)Cc1ccccc1-2 | 90.0 |
| 2 | c1ccc2c(c1)CCCC2 | 69.4 |
Save to disk for Chemprop CLI.
df_reg.to_csv("mp_data.csv", index=False)
len(df_reg), df_reg.head(2)
(575,
SMILES Melting Point
0 c1ccc2c(c1)CCOC2 65.8
1 c1ccc2c(c1)Cc1ccccc1-2 90.0)
Train a small model so it runs in class. We log common metrics.
!chemprop train \
--data-path mp_data.csv \
-t regression \
-s SMILES \
--target-columns "Melting Point" \
-o mp_model \
--num-replicates 1 \
--epochs 100 \
--save-smiles-splits \
--metrics mae rmse r2 \
--tracking-metric r2
2025-09-21T02:36:17 - INFO:chemprop.cli.main - Running in mode 'train' with args: {'smiles_columns': ['SMILES'], 'reaction_columns': None, 'no_header_row': False, 'num_workers': 0, 'batch_size': 64, 'accelerator': 'auto', 'devices': 'auto', 'rxn_mode': 'REAC_DIFF', 'multi_hot_atom_featurizer_mode': 'V2', 'keep_h': False, 'add_h': False, 'ignore_stereo': False, 'reorder_atoms': False, 'molecule_featurizers': None, 'descriptors_path': None, 'descriptors_columns': None, 'no_descriptor_scaling': False, 'no_atom_feature_scaling': False, 'no_atom_descriptor_scaling': False, 'no_bond_feature_scaling': False, 'no_bond_descriptor_scaling': False, 'atom_features_path': None, 'atom_descriptors_path': None, 'bond_features_path': None, 'bond_descriptors_path': None, 'constraints_path': None, 'constraints_to_targets': None, 'use_cuikmolmaker_featurization': False, 'config_path': None, 'data_path': PosixPath('mp_data.csv'), 'output_dir': PosixPath('mp_model'), 'remove_checkpoints': False, 'checkpoint': None, 'freeze_encoder': False, 'model_frzn': None, 'frzn_ffn_layers': 0, 'from_foundation': None, 'ensemble_size': 1, 'message_hidden_dim': 300, 'message_bias': False, 'depth': 3, 'undirected': False, 'dropout': 0.0, 'mpn_shared': False, 'aggregation': 'norm', 'aggregation_norm': 100, 'atom_messages': False, 'activation': 'RELU', 'activation_args': None, 'ffn_hidden_dim': 300, 'ffn_num_layers': 1, 'batch_norm': False, 'multiclass_num_classes': 3, 'atom_task_weights': None, 'atom_ffn_hidden_dim': 300, 'atom_ffn_num_layers': 1, 'atom_multiclass_num_classes': 3, 'bond_task_weights': None, 'bond_ffn_hidden_dim': 300, 'bond_ffn_num_layers': 1, 'bond_multiclass_num_classes': 3, 'atom_constrainer_ffn_hidden_dim': 300, 'atom_constrainer_ffn_num_layers': 1, 'bond_constrainer_ffn_hidden_dim': 300, 'bond_constrainer_ffn_num_layers': 1, 'weight_column': None, 'target_columns': ['Melting Point'], 'mol_target_columns': None, 'atom_target_columns': None, 'bond_target_columns': None, 'ignore_columns': None, 'no_cache': False, 'splits_column': None, 'task_type': 'regression', 'loss_function': None, 'v_kl': 0.0, 'eps': 1e-08, 'alpha': 0.1, 'metrics': ['mae', 'rmse', 'r2'], 'tracking_metric': 'r2', 'show_individual_scores': False, 'task_weights': None, 'warmup_epochs': 2, 'init_lr': 0.0001, 'max_lr': 0.001, 'final_lr': 0.0001, 'epochs': 100, 'patience': None, 'grad_clip': None, 'class_balance': False, 'split': 'RANDOM', 'split_sizes': [0.8, 0.1, 0.1], 'split_key_molecule': 0, 'num_replicates': 1, 'num_folds': None, 'save_smiles_splits': True, 'splits_file': None, 'data_seed': 0, 'pytorch_seed': None}
Wrote config file to mp_model/config.toml
2025-09-21T02:36:17 - INFO:chemprop.cli.train - Pulling data from file: mp_data.csv
2025-09-21T02:36:17 - WARNING:chemprop.data.splitting - The return type of make_split_indices has changed in v2.1 - see help(make_split_indices)
2025-09-21T02:36:17 - INFO:chemprop.cli.train - train/val/test split_0 sizes: [460, 57, 58]
2025-09-21T02:36:17 - INFO:chemprop.cli.train -
Summary of Training Data
┏━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━┓
┃ Statistic ┃ Value (Melting Point) ┃
┡━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━┩
│ Num. smiles │ 460 │
│ Num. targets │ 460 │
│ Num. NaN │ 0 │
│ Mean │ 133 │
│ Std. dev. │ 51.2 │
│ Median │ 129 │
│ % within 1 s.d. │ 75% │
│ % within 2 s.d. │ 96% │
└─────────────────┴───────────────────────┘
2025-09-21T02:36:17 - INFO:chemprop.cli.train -
Summary of Validation Data
┏━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━┓
┃ Statistic ┃ Value (Melting Point) ┃
┡━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━┩
│ Num. smiles │ 57 │
│ Num. targets │ 57 │
│ Num. NaN │ 0 │
│ Mean │ 123 │
│ Std. dev. │ 43.9 │
│ Median │ 124 │
│ % within 1 s.d. │ 67% │
│ % within 2 s.d. │ 96% │
└─────────────────┴───────────────────────┘
2025-09-21T02:36:17 - INFO:chemprop.cli.train -
Summary of Test Data
┏━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━┓
┃ Statistic ┃ Value (Melting Point) ┃
┡━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━┩
│ Num. smiles │ 58 │
│ Num. targets │ 58 │
│ Num. NaN │ 0 │
│ Mean │ 130 │
│ Std. dev. │ 50.1 │
│ Median │ 117 │
│ % within 1 s.d. │ 79% │
│ % within 2 s.d. │ 93% │
└─────────────────┴───────────────────────┘
2025-09-21T02:36:17 - INFO:chemprop.cli.train - Train data: mean = [133.13913042] | std = [51.15911504]
2025-09-21T02:36:17 - INFO:chemprop.cli.train - Caching training and validation datasets...
2025-09-21T02:36:18 - INFO:chemprop.cli.train - No loss function was specified! Using class default: <class 'chemprop.nn.metrics.MSE'>
2025-09-21T02:36:18 - INFO:chemprop.cli.train - MPNN(
(message_passing): BondMessagePassing(
(W_i): Linear(in_features=86, out_features=300, bias=False)
(W_h): Linear(in_features=300, out_features=300, bias=False)
(W_o): Linear(in_features=372, out_features=300, bias=True)
(dropout): Dropout(p=0.0, inplace=False)
(tau): ReLU()
(V_d_transform): Identity()
(graph_transform): GraphTransform(
(V_transform): Identity()
(E_transform): Identity()
)
)
(agg): NormAggregation()
(bn): Identity()
(predictor): RegressionFFN(
(ffn): MLP(
(0): Sequential(
(0): Linear(in_features=300, out_features=300, bias=True)
)
(1): Sequential(
(0): ReLU()
(1): Dropout(p=0.0, inplace=False)
(2): Linear(in_features=300, out_features=1, bias=True)
)
)
(criterion): MSE(task_weights=[[1.0]])
(output_transform): UnscaleTransform()
)
(X_d_transform): Identity()
(metrics): ModuleList(
(0): MAE(task_weights=[[1.0]])
(1): RMSE(task_weights=[[1.0]])
(2): R2Score()
(3): MSE(task_weights=[[1.0]])
)
)
GPU available: False, used: False
TPU available: False, using: 0 TPU cores
HPU available: False, using: 0 HPUs
2025-09-21 02:36:19.170674: E external/local_xla/xla/stream_executor/cuda/cuda_fft.cc:467] Unable to register cuFFT factory: Attempting to register factory for plugin cuFFT when one has already been registered
WARNING: All log messages before absl::InitializeLog() is called are written to STDERR
E0000 00:00:1758422179.210367 58576 cuda_dnn.cc:8579] Unable to register cuDNN factory: Attempting to register factory for plugin cuDNN when one has already been registered
E0000 00:00:1758422179.222474 58576 cuda_blas.cc:1407] Unable to register cuBLAS factory: Attempting to register factory for plugin cuBLAS when one has already been registered
W0000 00:00:1758422179.251528 58576 computation_placer.cc:177] computation placer already registered. Please check linkage and avoid linking the same target more than once.
W0000 00:00:1758422179.251595 58576 computation_placer.cc:177] computation placer already registered. Please check linkage and avoid linking the same target more than once.
W0000 00:00:1758422179.251605 58576 computation_placer.cc:177] computation placer already registered. Please check linkage and avoid linking the same target more than once.
W0000 00:00:1758422179.251613 58576 computation_placer.cc:177] computation placer already registered. Please check linkage and avoid linking the same target more than once.
2025-09-21 02:36:19.259832: I tensorflow/core/platform/cpu_feature_guard.cc:210] This TensorFlow binary is optimized to use available CPU instructions in performance-critical operations.
To enable the following instructions: AVX2 FMA, in other operations, rebuild TensorFlow with the appropriate compiler flags.
/usr/local/lib/python3.12/dist-packages/lightning/pytorch/callbacks/model_checkpoint.py:751: Checkpoint directory /content/mp_model/model_0/checkpoints exists and is not empty.
Loading `train_dataloader` to estimate number of stepping batches.
/usr/local/lib/python3.12/dist-packages/lightning/pytorch/loops/fit_loop.py:310: The number of training batches (8) is smaller than the logging interval Trainer(log_every_n_steps=50). Set a lower value for log_every_n_steps if you want to see logs for the training epoch.
| Name | Type | Params | Mode
---------------------------------------------------------------
0 | message_passing | BondMessagePassing | 227 K | train
1 | agg | NormAggregation | 0 | train
2 | bn | Identity | 0 | train
3 | predictor | RegressionFFN | 90.6 K | train
4 | X_d_transform | Identity | 0 | train
5 | metrics | ModuleList | 0 | train
---------------------------------------------------------------
318 K Trainable params
0 Non-trainable params
318 K Total params
1.273 Total estimated model params size (MB)
27 Modules in train mode
0 Modules in eval mode
/usr/local/lib/python3.12/dist-packages/lightning/pytorch/core/saving.py:363: Skipping 'metrics' parameter because it is not possible to safely dump to YAML.
Epoch 0: 100% 8/8 [00:01<00:00, 7.54it/s, v_num=3, train_loss_step=0.551]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.31it/s]
Epoch 1: 100% 8/8 [00:01<00:00, 7.80it/s, v_num=3, train_loss_step=1.150, val_loss=0.797, train_loss_epoch=0.986]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.24it/s]
Epoch 2: 100% 8/8 [00:01<00:00, 7.85it/s, v_num=3, train_loss_step=0.346, val_loss=0.800, train_loss_epoch=0.898]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.55it/s]
Epoch 3: 100% 8/8 [00:01<00:00, 7.86it/s, v_num=3, train_loss_step=0.496, val_loss=0.766, train_loss_epoch=0.821]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.17it/s]
Epoch 4: 100% 8/8 [00:01<00:00, 7.86it/s, v_num=3, train_loss_step=0.481, val_loss=0.651, train_loss_epoch=0.757]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.50it/s]
Epoch 5: 100% 8/8 [00:01<00:00, 7.83it/s, v_num=3, train_loss_step=0.368, val_loss=0.709, train_loss_epoch=0.718]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.00it/s]
Epoch 6: 100% 8/8 [00:01<00:00, 7.88it/s, v_num=3, train_loss_step=0.544, val_loss=0.540, train_loss_epoch=0.605]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.23it/s]
Epoch 7: 100% 8/8 [00:01<00:00, 5.59it/s, v_num=3, train_loss_step=0.953, val_loss=0.487, train_loss_epoch=0.582]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 6.82it/s]
Epoch 8: 100% 8/8 [00:01<00:00, 5.23it/s, v_num=3, train_loss_step=1.650, val_loss=0.631, train_loss_epoch=0.564]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 6.69it/s]
Epoch 9: 100% 8/8 [00:01<00:00, 7.39it/s, v_num=3, train_loss_step=1.360, val_loss=0.472, train_loss_epoch=0.494]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.47it/s]
Epoch 10: 100% 8/8 [00:01<00:00, 7.92it/s, v_num=3, train_loss_step=0.110, val_loss=0.411, train_loss_epoch=0.527]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 9.37it/s]
Epoch 11: 100% 8/8 [00:01<00:00, 7.77it/s, v_num=3, train_loss_step=0.214, val_loss=0.422, train_loss_epoch=0.424]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.64it/s]
Epoch 12: 100% 8/8 [00:01<00:00, 7.87it/s, v_num=3, train_loss_step=0.253, val_loss=0.436, train_loss_epoch=0.385]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.50it/s]
Epoch 13: 100% 8/8 [00:01<00:00, 7.80it/s, v_num=3, train_loss_step=0.470, val_loss=0.372, train_loss_epoch=0.371]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.56it/s]
Epoch 14: 100% 8/8 [00:01<00:00, 7.82it/s, v_num=3, train_loss_step=0.608, val_loss=0.323, train_loss_epoch=0.338]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.19it/s]
Epoch 15: 100% 8/8 [00:00<00:00, 8.01it/s, v_num=3, train_loss_step=0.187, val_loss=0.376, train_loss_epoch=0.326]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.43it/s]
Epoch 16: 100% 8/8 [00:01<00:00, 7.98it/s, v_num=3, train_loss_step=0.254, val_loss=0.310, train_loss_epoch=0.301]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.69it/s]
Epoch 17: 100% 8/8 [00:01<00:00, 6.99it/s, v_num=3, train_loss_step=0.292, val_loss=0.325, train_loss_epoch=0.288]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 6.84it/s]
Epoch 18: 100% 8/8 [00:01<00:00, 5.05it/s, v_num=3, train_loss_step=0.365, val_loss=0.292, train_loss_epoch=0.267]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 6.69it/s]
Epoch 19: 100% 8/8 [00:01<00:00, 6.34it/s, v_num=3, train_loss_step=0.197, val_loss=0.279, train_loss_epoch=0.246]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 9.83it/s]
Epoch 20: 100% 8/8 [00:01<00:00, 7.73it/s, v_num=3, train_loss_step=0.189, val_loss=0.255, train_loss_epoch=0.249]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 8.63it/s]
Epoch 21: 100% 8/8 [00:01<00:00, 7.54it/s, v_num=3, train_loss_step=0.263, val_loss=0.268, train_loss_epoch=0.249]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 9.76it/s]
Epoch 22: 100% 8/8 [00:01<00:00, 7.68it/s, v_num=3, train_loss_step=0.206, val_loss=0.375, train_loss_epoch=0.216]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.12it/s]
Epoch 23: 100% 8/8 [00:01<00:00, 7.78it/s, v_num=3, train_loss_step=0.177, val_loss=0.242, train_loss_epoch=0.235]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 9.89it/s]
Epoch 24: 100% 8/8 [00:01<00:00, 7.77it/s, v_num=3, train_loss_step=0.287, val_loss=0.232, train_loss_epoch=0.215]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.54it/s]
Epoch 25: 100% 8/8 [00:01<00:00, 7.97it/s, v_num=3, train_loss_step=0.0736, val_loss=0.227, train_loss_epoch=0.207]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.53it/s]
Epoch 26: 100% 8/8 [00:01<00:00, 7.84it/s, v_num=3, train_loss_step=0.166, val_loss=0.309, train_loss_epoch=0.226]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.26it/s]
Epoch 27: 100% 8/8 [00:00<00:00, 8.03it/s, v_num=3, train_loss_step=0.0895, val_loss=0.278, train_loss_epoch=0.197]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.52it/s]
Epoch 28: 100% 8/8 [00:01<00:00, 5.37it/s, v_num=3, train_loss_step=0.0522, val_loss=0.233, train_loss_epoch=0.194]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 6.79it/s]
Epoch 29: 100% 8/8 [00:01<00:00, 5.36it/s, v_num=3, train_loss_step=0.290, val_loss=0.224, train_loss_epoch=0.178]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.01it/s]
Epoch 30: 100% 8/8 [00:01<00:00, 7.82it/s, v_num=3, train_loss_step=0.250, val_loss=0.219, train_loss_epoch=0.173]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.35it/s]
Epoch 31: 100% 8/8 [00:01<00:00, 7.94it/s, v_num=3, train_loss_step=0.137, val_loss=0.240, train_loss_epoch=0.198]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 8.97it/s]
Epoch 32: 100% 8/8 [00:01<00:00, 7.92it/s, v_num=3, train_loss_step=0.247, val_loss=0.279, train_loss_epoch=0.186]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.40it/s]
Epoch 33: 100% 8/8 [00:01<00:00, 7.54it/s, v_num=3, train_loss_step=0.232, val_loss=0.240, train_loss_epoch=0.180]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.43it/s]
Epoch 34: 100% 8/8 [00:01<00:00, 7.82it/s, v_num=3, train_loss_step=0.217, val_loss=0.224, train_loss_epoch=0.166]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.15it/s]
Epoch 35: 100% 8/8 [00:00<00:00, 8.02it/s, v_num=3, train_loss_step=0.311, val_loss=0.256, train_loss_epoch=0.159]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.68it/s]
Epoch 36: 100% 8/8 [00:01<00:00, 7.78it/s, v_num=3, train_loss_step=0.201, val_loss=0.222, train_loss_epoch=0.160]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.57it/s]
Epoch 37: 100% 8/8 [00:01<00:00, 7.84it/s, v_num=3, train_loss_step=0.0628, val_loss=0.221, train_loss_epoch=0.163]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 9.96it/s]
Epoch 38: 100% 8/8 [00:01<00:00, 6.80it/s, v_num=3, train_loss_step=0.153, val_loss=0.237, train_loss_epoch=0.159]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 6.94it/s]
Epoch 39: 100% 8/8 [00:01<00:00, 5.27it/s, v_num=3, train_loss_step=0.191, val_loss=0.243, train_loss_epoch=0.164]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 6.81it/s]
Epoch 40: 100% 8/8 [00:01<00:00, 6.54it/s, v_num=3, train_loss_step=0.399, val_loss=0.248, train_loss_epoch=0.155]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.16it/s]
Epoch 41: 100% 8/8 [00:01<00:00, 7.94it/s, v_num=3, train_loss_step=0.155, val_loss=0.217, train_loss_epoch=0.165]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.60it/s]
Epoch 42: 100% 8/8 [00:01<00:00, 7.94it/s, v_num=3, train_loss_step=0.154, val_loss=0.218, train_loss_epoch=0.156]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.19it/s]
Epoch 43: 100% 8/8 [00:01<00:00, 7.97it/s, v_num=3, train_loss_step=0.226, val_loss=0.286, train_loss_epoch=0.156]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.61it/s]
Epoch 44: 100% 8/8 [00:01<00:00, 7.83it/s, v_num=3, train_loss_step=0.0866, val_loss=0.231, train_loss_epoch=0.165]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.02it/s]
Epoch 45: 100% 8/8 [00:01<00:00, 4.67it/s, v_num=3, train_loss_step=0.265, val_loss=0.221, train_loss_epoch=0.152]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.53it/s]
Epoch 46: 100% 8/8 [00:01<00:00, 7.95it/s, v_num=3, train_loss_step=0.0487, val_loss=0.243, train_loss_epoch=0.152]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 9.81it/s]
Epoch 47: 100% 8/8 [00:01<00:00, 7.97it/s, v_num=3, train_loss_step=0.170, val_loss=0.243, train_loss_epoch=0.149]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.81it/s]
Epoch 48: 100% 8/8 [00:01<00:00, 6.57it/s, v_num=3, train_loss_step=0.148, val_loss=0.234, train_loss_epoch=0.149]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 6.53it/s]
Epoch 49: 100% 8/8 [00:01<00:00, 5.32it/s, v_num=3, train_loss_step=0.117, val_loss=0.226, train_loss_epoch=0.143]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 6.84it/s]
Epoch 50: 100% 8/8 [00:01<00:00, 6.45it/s, v_num=3, train_loss_step=0.102, val_loss=0.258, train_loss_epoch=0.142]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 8.89it/s]
Epoch 51: 100% 8/8 [00:01<00:00, 7.94it/s, v_num=3, train_loss_step=0.132, val_loss=0.216, train_loss_epoch=0.150]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.65it/s]
Epoch 52: 100% 8/8 [00:01<00:00, 7.90it/s, v_num=3, train_loss_step=0.114, val_loss=0.220, train_loss_epoch=0.152]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.12it/s]
Epoch 53: 100% 8/8 [00:00<00:00, 8.01it/s, v_num=3, train_loss_step=0.115, val_loss=0.223, train_loss_epoch=0.144]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.47it/s]
Epoch 54: 100% 8/8 [00:01<00:00, 7.97it/s, v_num=3, train_loss_step=0.0449, val_loss=0.235, train_loss_epoch=0.143]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.75it/s]
Epoch 55: 100% 8/8 [00:01<00:00, 7.96it/s, v_num=3, train_loss_step=0.119, val_loss=0.230, train_loss_epoch=0.141]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.52it/s]
Epoch 56: 100% 8/8 [00:01<00:00, 7.80it/s, v_num=3, train_loss_step=0.156, val_loss=0.223, train_loss_epoch=0.140]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.10it/s]
Epoch 57: 100% 8/8 [00:01<00:00, 7.60it/s, v_num=3, train_loss_step=0.207, val_loss=0.226, train_loss_epoch=0.139]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.67it/s]
Epoch 58: 100% 8/8 [00:01<00:00, 7.96it/s, v_num=3, train_loss_step=0.154, val_loss=0.240, train_loss_epoch=0.140]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.65it/s]
Epoch 59: 100% 8/8 [00:01<00:00, 5.81it/s, v_num=3, train_loss_step=0.174, val_loss=0.216, train_loss_epoch=0.141]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 6.80it/s]
Epoch 60: 100% 8/8 [00:01<00:00, 5.44it/s, v_num=3, train_loss_step=0.168, val_loss=0.227, train_loss_epoch=0.138]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 6.79it/s]
Epoch 61: 100% 8/8 [00:01<00:00, 7.55it/s, v_num=3, train_loss_step=0.0818, val_loss=0.231, train_loss_epoch=0.138]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.01it/s]
Epoch 62: 100% 8/8 [00:01<00:00, 7.87it/s, v_num=3, train_loss_step=0.162, val_loss=0.229, train_loss_epoch=0.135]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.36it/s]
Epoch 63: 100% 8/8 [00:01<00:00, 7.87it/s, v_num=3, train_loss_step=0.0715, val_loss=0.218, train_loss_epoch=0.134]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 8.99it/s]
Epoch 64: 100% 8/8 [00:01<00:00, 7.79it/s, v_num=3, train_loss_step=0.128, val_loss=0.230, train_loss_epoch=0.136]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.58it/s]
Epoch 65: 100% 8/8 [00:01<00:00, 7.86it/s, v_num=3, train_loss_step=0.216, val_loss=0.225, train_loss_epoch=0.133]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.50it/s]
Epoch 66: 100% 8/8 [00:01<00:00, 7.93it/s, v_num=3, train_loss_step=0.118, val_loss=0.221, train_loss_epoch=0.134]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.64it/s]
Epoch 67: 100% 8/8 [00:00<00:00, 8.10it/s, v_num=3, train_loss_step=0.189, val_loss=0.237, train_loss_epoch=0.131]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.67it/s]
Epoch 68: 100% 8/8 [00:01<00:00, 7.99it/s, v_num=3, train_loss_step=0.161, val_loss=0.219, train_loss_epoch=0.132]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.46it/s]
Epoch 69: 100% 8/8 [00:01<00:00, 7.85it/s, v_num=3, train_loss_step=0.208, val_loss=0.241, train_loss_epoch=0.134]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 6.95it/s]
Epoch 70: 100% 8/8 [00:01<00:00, 5.36it/s, v_num=3, train_loss_step=0.119, val_loss=0.229, train_loss_epoch=0.131]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 6.54it/s]
Epoch 71: 100% 8/8 [00:01<00:00, 5.64it/s, v_num=3, train_loss_step=0.198, val_loss=0.222, train_loss_epoch=0.133]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.29it/s]
Epoch 72: 100% 8/8 [00:01<00:00, 7.83it/s, v_num=3, train_loss_step=0.117, val_loss=0.223, train_loss_epoch=0.132]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.26it/s]
Epoch 73: 100% 8/8 [00:01<00:00, 7.66it/s, v_num=3, train_loss_step=0.0715, val_loss=0.239, train_loss_epoch=0.133]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.25it/s]
Epoch 74: 100% 8/8 [00:01<00:00, 7.73it/s, v_num=3, train_loss_step=0.284, val_loss=0.219, train_loss_epoch=0.134]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.59it/s]
Epoch 75: 100% 8/8 [00:01<00:00, 7.69it/s, v_num=3, train_loss_step=0.135, val_loss=0.247, train_loss_epoch=0.130]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.28it/s]
Epoch 76: 100% 8/8 [00:01<00:00, 7.94it/s, v_num=3, train_loss_step=0.0589, val_loss=0.221, train_loss_epoch=0.133]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 8.96it/s]
Epoch 77: 100% 8/8 [00:01<00:00, 7.81it/s, v_num=3, train_loss_step=0.099, val_loss=0.235, train_loss_epoch=0.130]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.60it/s]
Epoch 78: 100% 8/8 [00:01<00:00, 7.97it/s, v_num=3, train_loss_step=0.0862, val_loss=0.223, train_loss_epoch=0.129]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.24it/s]
Epoch 79: 100% 8/8 [00:01<00:00, 7.99it/s, v_num=3, train_loss_step=0.116, val_loss=0.238, train_loss_epoch=0.127]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.19it/s]
Epoch 80: 100% 8/8 [00:01<00:00, 6.47it/s, v_num=3, train_loss_step=0.226, val_loss=0.222, train_loss_epoch=0.131]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 6.86it/s]
Epoch 81: 100% 8/8 [00:01<00:00, 5.25it/s, v_num=3, train_loss_step=0.135, val_loss=0.228, train_loss_epoch=0.128]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 6.87it/s]
Epoch 82: 100% 8/8 [00:01<00:00, 6.01it/s, v_num=3, train_loss_step=0.110, val_loss=0.240, train_loss_epoch=0.130]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.07it/s]
Epoch 83: 100% 8/8 [00:01<00:00, 7.58it/s, v_num=3, train_loss_step=0.0917, val_loss=0.221, train_loss_epoch=0.129]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.35it/s]
Epoch 84: 100% 8/8 [00:01<00:00, 7.70it/s, v_num=3, train_loss_step=0.114, val_loss=0.227, train_loss_epoch=0.127]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.20it/s]
Epoch 85: 100% 8/8 [00:01<00:00, 7.61it/s, v_num=3, train_loss_step=0.134, val_loss=0.223, train_loss_epoch=0.126]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.63it/s]
Epoch 86: 100% 8/8 [00:01<00:00, 7.65it/s, v_num=3, train_loss_step=0.156, val_loss=0.232, train_loss_epoch=0.127]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.30it/s]
Epoch 87: 100% 8/8 [00:01<00:00, 7.77it/s, v_num=3, train_loss_step=0.125, val_loss=0.222, train_loss_epoch=0.127]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 9.05it/s]
Epoch 88: 100% 8/8 [00:01<00:00, 7.73it/s, v_num=3, train_loss_step=0.142, val_loss=0.233, train_loss_epoch=0.126]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.49it/s]
Epoch 89: 100% 8/8 [00:01<00:00, 7.74it/s, v_num=3, train_loss_step=0.0796, val_loss=0.219, train_loss_epoch=0.128]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.05it/s]
Epoch 90: 100% 8/8 [00:01<00:00, 7.66it/s, v_num=3, train_loss_step=0.0883, val_loss=0.227, train_loss_epoch=0.127]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 9.95it/s]
Epoch 91: 100% 8/8 [00:01<00:00, 5.46it/s, v_num=3, train_loss_step=0.150, val_loss=0.226, train_loss_epoch=0.126]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 6.83it/s]
Epoch 92: 100% 8/8 [00:01<00:00, 5.16it/s, v_num=3, train_loss_step=0.133, val_loss=0.220, train_loss_epoch=0.127]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 6.57it/s]
Epoch 93: 100% 8/8 [00:01<00:00, 7.77it/s, v_num=3, train_loss_step=0.110, val_loss=0.230, train_loss_epoch=0.130]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.47it/s]
Epoch 94: 100% 8/8 [00:01<00:00, 7.67it/s, v_num=3, train_loss_step=0.0656, val_loss=0.227, train_loss_epoch=0.125]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.26it/s]
Epoch 95: 100% 8/8 [00:01<00:00, 7.66it/s, v_num=3, train_loss_step=0.134, val_loss=0.224, train_loss_epoch=0.125]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.46it/s]
Epoch 96: 100% 8/8 [00:01<00:00, 6.60it/s, v_num=3, train_loss_step=0.106, val_loss=0.231, train_loss_epoch=0.124]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 9.54it/s]
Epoch 97: 100% 8/8 [00:01<00:00, 7.69it/s, v_num=3, train_loss_step=0.123, val_loss=0.225, train_loss_epoch=0.127]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.87it/s]
Epoch 98: 100% 8/8 [00:01<00:00, 7.61it/s, v_num=3, train_loss_step=0.141, val_loss=0.228, train_loss_epoch=0.126]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.18it/s]
Epoch 99: 100% 8/8 [00:01<00:00, 7.69it/s, v_num=3, train_loss_step=0.170, val_loss=0.226, train_loss_epoch=0.126]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.26it/s]
Epoch 99: 100% 8/8 [00:01<00:00, 6.98it/s, v_num=3, train_loss_step=0.170, val_loss=0.228, train_loss_epoch=0.130]`Trainer.fit` stopped: `max_epochs=100` reached.
Epoch 99: 100% 8/8 [00:01<00:00, 6.88it/s, v_num=3, train_loss_step=0.170, val_loss=0.228, train_loss_epoch=0.130]
/usr/local/lib/python3.12/dist-packages/lightning/pytorch/trainer/connectors/checkpoint_connector.py:149: `.predict(ckpt_path=None)` was called without a model. The best model of the previous `fit` call will be used. You can pass `.predict(ckpt_path='best')` to use the best model or `.predict(ckpt_path='last')` to use the last model. If you pass a value, this warning will be silenced.
Restoring states from the checkpoint path at /content/mp_model/model_0/checkpoints/best-epoch=50-val_r2=0.71.ckpt
Loaded model weights from the checkpoint at /content/mp_model/model_0/checkpoints/best-epoch=50-val_r2=0.71.ckpt
Predicting DataLoader 0: 100% 1/1 [00:00<00:00, 19.21it/s]
2025-09-21T02:38:28 - INFO:chemprop.cli.train - Test Set results:
2025-09-21T02:38:28 - INFO:chemprop.cli.train - test/mae: 16.6578369140625
2025-09-21T02:38:28 - INFO:chemprop.cli.train - test/rmse: 20.868921279907227
2025-09-21T02:38:28 - INFO:chemprop.cli.train - test/r2: 0.826757027488992
2025-09-21T02:38:28 - INFO:chemprop.cli.train - Best model saved to 'mp_model/model_0/best.pt'
Make quick predictions on a few molecules.
smiles_list = [
"CO",
"c1ccc2c(c1)CCCC2",
"CCCCCCCCC(=O)O",
"CCN(CC)CC"
]
pd.DataFrame({"SMILES": smiles_list}).to_csv("custom_smiles_reg.csv", index=False)
!chemprop predict \
--test-path custom_smiles_reg.csv \
--model-paths mp_model/model_0/best.pt \
--preds-path mp_preds.csv
pd.read_csv("mp_preds.csv")
2025-09-21T02:38:41 - INFO:chemprop.cli.main - Running in mode 'predict' with args: {'smiles_columns': None, 'reaction_columns': None, 'no_header_row': False, 'num_workers': 0, 'batch_size': 64, 'accelerator': 'auto', 'devices': 'auto', 'rxn_mode': 'REAC_DIFF', 'multi_hot_atom_featurizer_mode': 'V2', 'keep_h': False, 'add_h': False, 'ignore_stereo': False, 'reorder_atoms': False, 'molecule_featurizers': None, 'descriptors_path': None, 'descriptors_columns': None, 'no_descriptor_scaling': False, 'no_atom_feature_scaling': False, 'no_atom_descriptor_scaling': False, 'no_bond_feature_scaling': False, 'no_bond_descriptor_scaling': False, 'atom_features_path': None, 'atom_descriptors_path': None, 'bond_features_path': None, 'bond_descriptors_path': None, 'constraints_path': None, 'constraints_to_targets': None, 'use_cuikmolmaker_featurization': False, 'test_path': PosixPath('custom_smiles_reg.csv'), 'output': PosixPath('mp_preds.csv'), 'drop_extra_columns': False, 'model_paths': [PosixPath('mp_model/model_0/best.pt')], 'cal_path': None, 'uncertainty_method': 'none', 'calibration_method': None, 'evaluation_methods': None, 'uncertainty_dropout_p': 0.1, 'dropout_sampling_size': 10, 'calibration_interval_percentile': 95, 'conformal_alpha': 0.1, 'cal_descriptors_path': None, 'cal_atom_features_path': None, 'cal_atom_descriptors_path': None, 'cal_bond_features_path': None, 'cal_bond_descriptors_path': None, 'cal_constraints_path': None}
2025-09-21T02:38:41 - INFO:chemprop.cli.predict - test size: 4
💡 Tip: For seamless cloud uploads and versioning, try installing [litmodels](https://pypi.org/project/litmodels/) to enable LitModelCheckpoint, which syncs automatically with the Lightning model registry.
GPU available: False, used: False
TPU available: False, using: 0 TPU cores
HPU available: False, using: 0 HPUs
Predicting: | | 0/? [00:00<?, ?it/s]
Predicting: | | 0/? [00:00<?, ?it/s]
Predicting DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Predicting DataLoader 0: 100% 1/1 [00:00<00:00, 239.83it/s]
Predicting DataLoader 0: 100% 1/1 [00:00<00:00, 219.64it/s]
2025-09-21T02:38:41 - INFO:chemprop.cli.predict - Predictions saved to 'mp_preds.csv'
| SMILES | Melting Point | |
|---|---|---|
| 0 | CO | 76.238464 |
| 1 | c1ccc2c(c1)CCCC2 | 76.600280 |
| 2 | CCCCCCCCC(=O)O | 91.757965 |
| 3 | CCN(CC)CC | 80.074980 |
7.3 Reactivity classification (C–H oxidation dataset)#
We use the Reactivity column and convert it to binary 0/1.
df = pd.read_csv(url)
df["Reactivity_bin"] = df["Reactivity"].replace({-1: 0}).astype(int)
df[["SMILES","Reactivity","Reactivity_bin"]].head(3)
| SMILES | Reactivity | Reactivity_bin | |
|---|---|---|---|
| 0 | c1ccc2c(c1)CCOC2 | 1 | 1 |
| 1 | c1ccc2c(c1)Cc1ccccc1-2 | 1 | 1 |
| 2 | c1ccc2c(c1)CCCC2 | 1 | 1 |
Write a minimal file.
df[["SMILES", "Reactivity_bin"]].to_csv("reactivity_data_bin.csv", index=False)
# Optional: sanity check the class balance
print(df["Reactivity"].value_counts(dropna=False).to_dict())
print(df["Reactivity_bin"].value_counts(dropna=False).to_dict())
{-1: 311, 1: 264}
{0: 311, 1: 264}
Train a short classification model.
!chemprop train \
--data-path reactivity_data_bin.csv \
-t classification \
-s SMILES \
--target-columns Reactivity_bin \
-o reactivity_model \
--num-replicates 1 \
--epochs 15 \
--class-balance \
--metrics roc prc accuracy \
--tracking-metric roc
2025-09-21T02:30:14 - INFO:chemprop.cli.main - Running in mode 'train' with args: {'smiles_columns': ['SMILES'], 'reaction_columns': None, 'no_header_row': False, 'num_workers': 0, 'batch_size': 64, 'accelerator': 'auto', 'devices': 'auto', 'rxn_mode': 'REAC_DIFF', 'multi_hot_atom_featurizer_mode': 'V2', 'keep_h': False, 'add_h': False, 'ignore_stereo': False, 'reorder_atoms': False, 'molecule_featurizers': None, 'descriptors_path': None, 'descriptors_columns': None, 'no_descriptor_scaling': False, 'no_atom_feature_scaling': False, 'no_atom_descriptor_scaling': False, 'no_bond_feature_scaling': False, 'no_bond_descriptor_scaling': False, 'atom_features_path': None, 'atom_descriptors_path': None, 'bond_features_path': None, 'bond_descriptors_path': None, 'constraints_path': None, 'constraints_to_targets': None, 'use_cuikmolmaker_featurization': False, 'config_path': None, 'data_path': PosixPath('reactivity_data_bin.csv'), 'output_dir': PosixPath('reactivity_model'), 'remove_checkpoints': False, 'checkpoint': None, 'freeze_encoder': False, 'model_frzn': None, 'frzn_ffn_layers': 0, 'from_foundation': None, 'ensemble_size': 1, 'message_hidden_dim': 300, 'message_bias': False, 'depth': 3, 'undirected': False, 'dropout': 0.0, 'mpn_shared': False, 'aggregation': 'norm', 'aggregation_norm': 100, 'atom_messages': False, 'activation': 'RELU', 'activation_args': None, 'ffn_hidden_dim': 300, 'ffn_num_layers': 1, 'batch_norm': False, 'multiclass_num_classes': 3, 'atom_task_weights': None, 'atom_ffn_hidden_dim': 300, 'atom_ffn_num_layers': 1, 'atom_multiclass_num_classes': 3, 'bond_task_weights': None, 'bond_ffn_hidden_dim': 300, 'bond_ffn_num_layers': 1, 'bond_multiclass_num_classes': 3, 'atom_constrainer_ffn_hidden_dim': 300, 'atom_constrainer_ffn_num_layers': 1, 'bond_constrainer_ffn_hidden_dim': 300, 'bond_constrainer_ffn_num_layers': 1, 'weight_column': None, 'target_columns': ['Reactivity_bin'], 'mol_target_columns': None, 'atom_target_columns': None, 'bond_target_columns': None, 'ignore_columns': None, 'no_cache': False, 'splits_column': None, 'task_type': 'classification', 'loss_function': None, 'v_kl': 0.0, 'eps': 1e-08, 'alpha': 0.1, 'metrics': ['roc', 'prc', 'accuracy'], 'tracking_metric': 'roc', 'show_individual_scores': False, 'task_weights': None, 'warmup_epochs': 2, 'init_lr': 0.0001, 'max_lr': 0.001, 'final_lr': 0.0001, 'epochs': 15, 'patience': None, 'grad_clip': None, 'class_balance': True, 'split': 'RANDOM', 'split_sizes': [0.8, 0.1, 0.1], 'split_key_molecule': 0, 'num_replicates': 1, 'num_folds': None, 'save_smiles_splits': False, 'splits_file': None, 'data_seed': 0, 'pytorch_seed': None}
Wrote config file to reactivity_model/config.toml
2025-09-21T02:30:14 - INFO:chemprop.cli.train - Pulling data from file: reactivity_data_bin.csv
2025-09-21T02:30:15 - WARNING:chemprop.data.splitting - The return type of make_split_indices has changed in v2.1 - see help(make_split_indices)
2025-09-21T02:30:15 - INFO:chemprop.cli.train - train/val/test split_0 sizes: [460, 57, 58]
2025-09-21T02:30:15 - INFO:chemprop.cli.train -
Summary of Training Data
┏━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┓
┃ Class ┃ Count/Percent Reactivity_bin ┃
┡━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┩
│ 0.0 │ 255/55% │
│ 1.0 │ 205/45% │
│ NaN │ 0/0% │
│ Total │ 460/100.0% │
└───────┴──────────────────────────────┘
2025-09-21T02:30:15 - INFO:chemprop.cli.train -
Summary of Validation Data
┏━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┓
┃ Class ┃ Count/Percent Reactivity_bin ┃
┡━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┩
│ 0.0 │ 29/51% │
│ 1.0 │ 28/49% │
│ NaN │ 0/0% │
│ Total │ 57/100.0% │
└───────┴──────────────────────────────┘
2025-09-21T02:30:15 - INFO:chemprop.cli.train -
Summary of Test Data
┏━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┓
┃ Class ┃ Count/Percent Reactivity_bin ┃
┡━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┩
│ 0.0 │ 27/47% │
│ 1.0 │ 31/53% │
│ NaN │ 0/0% │
│ Total │ 58/100.0% │
└───────┴──────────────────────────────┘
2025-09-21T02:30:15 - INFO:chemprop.cli.train - Caching training and validation datasets...
2025-09-21T02:30:15 - INFO:chemprop.cli.train - No loss function was specified! Using class default: <class 'chemprop.nn.metrics.BCELoss'>
2025-09-21T02:30:15 - INFO:chemprop.cli.train - MPNN(
(message_passing): BondMessagePassing(
(W_i): Linear(in_features=86, out_features=300, bias=False)
(W_h): Linear(in_features=300, out_features=300, bias=False)
(W_o): Linear(in_features=372, out_features=300, bias=True)
(dropout): Dropout(p=0.0, inplace=False)
(tau): ReLU()
(V_d_transform): Identity()
(graph_transform): GraphTransform(
(V_transform): Identity()
(E_transform): Identity()
)
)
(agg): NormAggregation()
(bn): Identity()
(predictor): BinaryClassificationFFN(
(ffn): MLP(
(0): Sequential(
(0): Linear(in_features=300, out_features=300, bias=True)
)
(1): Sequential(
(0): ReLU()
(1): Dropout(p=0.0, inplace=False)
(2): Linear(in_features=300, out_features=1, bias=True)
)
)
(criterion): BCELoss(task_weights=[[1.0]])
(output_transform): Identity()
)
(X_d_transform): Identity()
(metrics): ModuleList(
(0): BinaryAUROC()
(1): BinaryAUPRC()
(2): BinaryAccuracy()
(3): BCELoss(task_weights=[[1.0]])
)
)
GPU available: False, used: False
TPU available: False, using: 0 TPU cores
HPU available: False, using: 0 HPUs
2025-09-21 02:30:15.974383: E external/local_xla/xla/stream_executor/cuda/cuda_fft.cc:467] Unable to register cuFFT factory: Attempting to register factory for plugin cuFFT when one has already been registered
WARNING: All log messages before absl::InitializeLog() is called are written to STDERR
E0000 00:00:1758421816.001418 57061 cuda_dnn.cc:8579] Unable to register cuDNN factory: Attempting to register factory for plugin cuDNN when one has already been registered
E0000 00:00:1758421816.009690 57061 cuda_blas.cc:1407] Unable to register cuBLAS factory: Attempting to register factory for plugin cuBLAS when one has already been registered
W0000 00:00:1758421816.031522 57061 computation_placer.cc:177] computation placer already registered. Please check linkage and avoid linking the same target more than once.
W0000 00:00:1758421816.031593 57061 computation_placer.cc:177] computation placer already registered. Please check linkage and avoid linking the same target more than once.
W0000 00:00:1758421816.031602 57061 computation_placer.cc:177] computation placer already registered. Please check linkage and avoid linking the same target more than once.
W0000 00:00:1758421816.031610 57061 computation_placer.cc:177] computation placer already registered. Please check linkage and avoid linking the same target more than once.
2025-09-21 02:30:16.037344: I tensorflow/core/platform/cpu_feature_guard.cc:210] This TensorFlow binary is optimized to use available CPU instructions in performance-critical operations.
To enable the following instructions: AVX2 FMA, in other operations, rebuild TensorFlow with the appropriate compiler flags.
Loading `train_dataloader` to estimate number of stepping batches.
/usr/local/lib/python3.12/dist-packages/lightning/pytorch/loops/fit_loop.py:310: The number of training batches (7) is smaller than the logging interval Trainer(log_every_n_steps=50). Set a lower value for log_every_n_steps if you want to see logs for the training epoch.
| Name | Type | Params | Mode
--------------------------------------------------------------------
0 | message_passing | BondMessagePassing | 227 K | train
1 | agg | NormAggregation | 0 | train
2 | bn | Identity | 0 | train
3 | predictor | BinaryClassificationFFN | 90.6 K | train
4 | X_d_transform | Identity | 0 | train
5 | metrics | ModuleList | 0 | train
--------------------------------------------------------------------
318 K Trainable params
0 Non-trainable params
318 K Total params
1.273 Total estimated model params size (MB)
27 Modules in train mode
0 Modules in eval mode
/usr/local/lib/python3.12/dist-packages/lightning/pytorch/core/saving.py:363: Skipping 'metrics' parameter because it is not possible to safely dump to YAML.
Epoch 0: 100% 7/7 [00:00<00:00, 7.31it/s, v_num=0, train_loss_step=0.687]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 10.21it/s]
Epoch 1: 100% 7/7 [00:00<00:00, 7.63it/s, v_num=0, train_loss_step=0.595, val_loss=0.685, train_loss_epoch=0.689]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 9.91it/s]
Epoch 2: 100% 7/7 [00:01<00:00, 5.31it/s, v_num=0, train_loss_step=0.512, val_loss=0.631, train_loss_epoch=0.656]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 6.38it/s]
Epoch 3: 100% 7/7 [00:01<00:00, 4.93it/s, v_num=0, train_loss_step=0.473, val_loss=0.595, train_loss_epoch=0.536]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 7.90it/s]
Epoch 4: 100% 7/7 [00:01<00:00, 5.34it/s, v_num=0, train_loss_step=0.433, val_loss=0.603, train_loss_epoch=0.504]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 6.65it/s]
Epoch 5: 100% 7/7 [00:01<00:00, 4.29it/s, v_num=0, train_loss_step=0.266, val_loss=0.573, train_loss_epoch=0.443]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 4.32it/s]
Epoch 6: 100% 7/7 [00:01<00:00, 4.99it/s, v_num=0, train_loss_step=0.408, val_loss=0.532, train_loss_epoch=0.434]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 5.97it/s]
Epoch 7: 100% 7/7 [00:01<00:00, 5.55it/s, v_num=0, train_loss_step=0.652, val_loss=0.526, train_loss_epoch=0.405]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 6.96it/s]
Epoch 8: 100% 7/7 [00:01<00:00, 5.01it/s, v_num=0, train_loss_step=0.306, val_loss=0.515, train_loss_epoch=0.387]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 6.13it/s]
Epoch 9: 100% 7/7 [00:01<00:00, 6.60it/s, v_num=0, train_loss_step=0.436, val_loss=0.496, train_loss_epoch=0.373]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 9.06it/s]
Epoch 10: 100% 7/7 [00:00<00:00, 7.78it/s, v_num=0, train_loss_step=0.247, val_loss=0.479, train_loss_epoch=0.367]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 8.93it/s]
Epoch 11: 100% 7/7 [00:00<00:00, 7.81it/s, v_num=0, train_loss_step=0.436, val_loss=0.472, train_loss_epoch=0.358]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 9.07it/s]
Epoch 12: 100% 7/7 [00:00<00:00, 7.78it/s, v_num=0, train_loss_step=0.291, val_loss=0.461, train_loss_epoch=0.349]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 8.84it/s]
Epoch 13: 100% 7/7 [00:00<00:00, 7.48it/s, v_num=0, train_loss_step=0.295, val_loss=0.454, train_loss_epoch=0.343]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 6.81it/s]
Epoch 14: 100% 7/7 [00:01<00:00, 4.44it/s, v_num=0, train_loss_step=0.367, val_loss=0.448, train_loss_epoch=0.339]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: | | 0/? [00:00<?, ?it/s]
Validation DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Validation DataLoader 0: 100% 1/1 [00:00<00:00, 5.01it/s]
Epoch 14: 100% 7/7 [00:01<00:00, 3.89it/s, v_num=0, train_loss_step=0.367, val_loss=0.439, train_loss_epoch=0.329]`Trainer.fit` stopped: `max_epochs=15` reached.
Epoch 14: 100% 7/7 [00:01<00:00, 3.79it/s, v_num=0, train_loss_step=0.367, val_loss=0.439, train_loss_epoch=0.329]
/usr/local/lib/python3.12/dist-packages/lightning/pytorch/trainer/connectors/checkpoint_connector.py:149: `.predict(ckpt_path=None)` was called without a model. The best model of the previous `fit` call will be used. You can pass `.predict(ckpt_path='best')` to use the best model or `.predict(ckpt_path='last')` to use the last model. If you pass a value, this warning will be silenced.
Restoring states from the checkpoint path at /content/reactivity_model/model_0/checkpoints/best-epoch=14-val_roc=0.86.ckpt
Loaded model weights from the checkpoint at /content/reactivity_model/model_0/checkpoints/best-epoch=14-val_roc=0.86.ckpt
Predicting DataLoader 0: 100% 1/1 [00:00<00:00, 7.23it/s]
2025-09-21T02:30:40 - INFO:chemprop.cli.train - Test Set results:
2025-09-21T02:30:40 - INFO:chemprop.cli.train - test/roc: 0.9510154724121094
2025-09-21T02:30:40 - INFO:chemprop.cli.train - test/prc: 0.9459617137908936
2025-09-21T02:30:40 - INFO:chemprop.cli.train - test/accuracy: 0.8793103694915771
2025-09-21T02:30:41 - INFO:chemprop.cli.train - Best model saved to 'reactivity_model/model_0/best.pt'
Predict on new SMILES.
smiles_list = [
"CCO",
"c1ccccc1C(F)",
"C1=C([C@@H]2C[C@H](C1)C2(C)C)",
"C1=CC=CC=C1C=O",
"CCN(CC)CC",
"c1cccc(C=CC)c1"
]
pd.DataFrame({"SMILES": smiles_list}).to_csv("custom_smiles.csv", index=False)
!chemprop predict \
--test-path custom_smiles.csv \
--model-paths reactivity_model/model_0/best.pt \
--preds-path custom_preds.csv
pd.read_csv("custom_preds.csv")
2025-09-21T02:32:16 - INFO:chemprop.cli.main - Running in mode 'predict' with args: {'smiles_columns': None, 'reaction_columns': None, 'no_header_row': False, 'num_workers': 0, 'batch_size': 64, 'accelerator': 'auto', 'devices': 'auto', 'rxn_mode': 'REAC_DIFF', 'multi_hot_atom_featurizer_mode': 'V2', 'keep_h': False, 'add_h': False, 'ignore_stereo': False, 'reorder_atoms': False, 'molecule_featurizers': None, 'descriptors_path': None, 'descriptors_columns': None, 'no_descriptor_scaling': False, 'no_atom_feature_scaling': False, 'no_atom_descriptor_scaling': False, 'no_bond_feature_scaling': False, 'no_bond_descriptor_scaling': False, 'atom_features_path': None, 'atom_descriptors_path': None, 'bond_features_path': None, 'bond_descriptors_path': None, 'constraints_path': None, 'constraints_to_targets': None, 'use_cuikmolmaker_featurization': False, 'test_path': PosixPath('custom_smiles.csv'), 'output': PosixPath('custom_preds.csv'), 'drop_extra_columns': False, 'model_paths': [PosixPath('reactivity_model/model_0/best.pt')], 'cal_path': None, 'uncertainty_method': 'none', 'calibration_method': None, 'evaluation_methods': None, 'uncertainty_dropout_p': 0.1, 'dropout_sampling_size': 10, 'calibration_interval_percentile': 95, 'conformal_alpha': 0.1, 'cal_descriptors_path': None, 'cal_atom_features_path': None, 'cal_atom_descriptors_path': None, 'cal_bond_features_path': None, 'cal_bond_descriptors_path': None, 'cal_constraints_path': None}
2025-09-21T02:32:16 - INFO:chemprop.cli.predict - test size: 6
💡 Tip: For seamless cloud uploads and versioning, try installing [litmodels](https://pypi.org/project/litmodels/) to enable LitModelCheckpoint, which syncs automatically with the Lightning model registry.
GPU available: False, used: False
TPU available: False, using: 0 TPU cores
HPU available: False, using: 0 HPUs
Predicting: | | 0/? [00:00<?, ?it/s]
Predicting: | | 0/? [00:00<?, ?it/s]
Predicting DataLoader 0: 0% 0/1 [00:00<?, ?it/s]
Predicting DataLoader 0: 100% 1/1 [00:00<00:00, 206.79it/s]
Predicting DataLoader 0: 100% 1/1 [00:00<00:00, 192.29it/s]
2025-09-21T02:32:16 - INFO:chemprop.cli.predict - Predictions saved to 'custom_preds.csv'
| SMILES | Reactivity_bin | |
|---|---|---|
| 0 | CCO | 0.588819 |
| 1 | c1ccccc1C(F) | 0.690307 |
| 2 | C1=C([C@@H]2C[C@H](C1)C2(C)C) | 0.899083 |
| 3 | C1=CC=CC=C1C=O | 0.461047 |
| 4 | CCN(CC)CC | 0.745579 |
| 5 | c1cccc(C=CC)c1 | 0.628841 |
7. Quick Reference#
An MLP ignores graph structure. A GNN uses edges to mix neighbor information.
The core update is neighbor aggregation followed by a learnable transformation.
Chemprop encodes many best practices so you can focus on data and targets.
8. Glossary#
graph neural network
A neural model that updates node states by aggregating messages from neighbors.
message passing
The update step where a node combines its own vector with aggregated neighbor vectors.
readout (pooling)
Operation that compresses node states into a single graph vector, often by sum, mean, or max.
edge index
A 2×E tensor listing source and destination of each edge.
Chemprop
A practical library that trains message passing networks directly from SMILES.
replicate
Independent training run with a different random seed. Often ensembled for stability.
tracking metric
The metric used to pick the best checkpoint during training.
class balance
Loss weighting that compensates for skewed class proportions.
10. In‑class activity#
Q1. Chemprop regression on melting point#
Train Chemprop on
mp_data.csvfor--epochs 20Predict on at least 5 new SMILES of your choice and list the predictions
# TO DO
Q2. Chemprop classification on toxicity (student challenge)#
Convert
Toxicityto1/0using the mapping{toxic:1, non_toxic:0}Save
["SMILES","Toxicity_bin"]totox_data.csvTrain with
--class-balance --epochs 20and metricsroc prc accuracyPredict on a small set of SMILES;
Optional: show the class probability
# TO DO
Solution Q1:#
df_reg = df[["SMILES","Melting Point"]].dropna().copy()
df_reg.to_csv("mp_data.csv", index=False)
!chemprop train \
--data-path mp_data.csv \
-t regression \
-s SMILES \
--target-columns "Melting Point" \
-o mp_model_q4 \
--num-replicates 1 \
--epochs 20 \
--metrics mae rmse r2 \
--tracking-metric r2
pd.DataFrame({"SMILES": ["CCO","c1ccccc1","CC(=O)O","CCN(CC)CC","O=C(O)C(O)C"]}).to_csv("q4_smiles.csv", index=False)
!chemprop predict \
--test-path q4_smiles.csv \
--model-paths mp_model_q4/replicate_0/model_0/best.pt \
--preds-path q4_preds.csv
pd.read_csv("q4_preds.csv")
Solution Q2#
df = pd.read_csv(url)
df = df[["SMILES","Toxicity"]].dropna().copy()
df["Toxicity_bin"] = df["Toxicity"].str.lower().map({"toxic":1, "non_toxic":0}).astype(int)
df[["SMILES","Toxicity_bin"]].to_csv("tox_data.csv", index=False)
!chemprop train \
--data-path tox_data.csv \
-t classification \
-s SMILES \
--target-columns Toxicity_bin \
-o tox_model \
--num-replicates 1 \
--epochs 20 \
--class-balance \
--metrics roc prc accuracy \
--tracking-metric roc
pd.DataFrame({"SMILES": ["CCO","c1ccccc1","O=[N+](=O)[O-]","ClCCl","CC(=O)Cl"]}).to_csv("q5_smiles.csv", index=False)
!chemprop predict \
--test-path q5_smiles.csv \
--model-paths tox_model/replicate_0/model_0/best.pt \
--preds-path q5_preds.csv
pd.read_csv("q5_preds.csv")

