Lecture 8 - Neural Networks#
Learning goals#
Explain a simple multilayer perceptron: layers, weights, bias, activation, loss, optimizer.
Build your first neural network for a toy problem, then for small chemistry tasks.
Train
MLPRegressorandMLPClassifierstep by step with diagnostics.Read training curves, avoid overfitting.
0. Setup#
We will stick to a light stack so it runs everywhere.
1. What is a neural network#
Picture in words
A multilayer perceptron (MLP) takes an input vector x, multiplies by a weight matrix, adds a bias, applies a nonlinear activation like ReLU or tanh, then repeats for one or more hidden layers, and finally outputs a number (regression) or class score (classification).
Key pieces:
Weights
Wand biasbset how features combine.Activation makes the model nonlinear so it can bend decision boundaries.
Loss measures error. We update weights to reduce loss on the training set.
Optimizer uses gradients to move weights a little bit each iteration (epoch).
We start tiny with a simulated dataset so we can see each piece clearly.
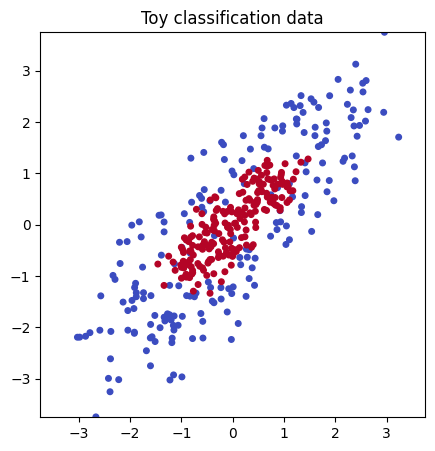
1.1 Generate a toy dataset and plot#
rng = np.random.RandomState(0)
n = 200
# --- Base donut in 2D ---
# Outer ring
theta = rng.uniform(0, 2*np.pi, size=n)
r = 1.0 + 0.3*rng.randn(n)
X1 = np.c_[3 * r*np.cos(theta), r*np.sin(theta)] # stretch x by 3
y1 = np.zeros(n, dtype=int)
# Inner ring
theta2 = rng.uniform(0, 2*np.pi, size=n)
r2 = 0.4 + 0.15*rng.randn(n)
X2 = np.c_[3 * r2*np.cos(theta2), r2*np.sin(theta2)] # stretch x by 3
y2 = np.ones(n, dtype=int)
# Combine 2D
X2d = np.vstack([X1, X2])
y_toy = np.hstack([y1, y2])
angle = np.pi / 4
R = np.array([[np.cos(angle), -np.sin(angle)],
[np.sin(angle), np.cos(angle)]])
X2d = X2d @ R.T
f_big = rng.normal(0, 10000.0, size=(X2d.shape[0], 1))
f_small = rng.normal(0, 0.0001, size=(X2d.shape[0], 1))
f_mid = rng.normal(0, 3.0, size=(X2d.shape[0], 1))
X_toy = np.hstack([X2d, f_big, f_small, f_mid])
# --- Plot using the first two columns only, keep square view ---
plt.figure(figsize=(5,5))
plt.scatter(X_toy[:,0], X_toy[:,1], c=y_toy, cmap="coolwarm", s=16)
lim = max(abs(X_toy[:,0]).max(), abs(X_toy[:,1]).max())
plt.xlim(-lim, lim)
plt.ylim(-lim, lim)
plt.gca().set_aspect("equal")
plt.title("Toy classification data")
plt.show()
1.2 Split and try a linear model#
Why a baseline
As in past lectures, a simple baseline tells us if a complex model is truly needed.
In scikit-learn, a pipeline is a convenient way to chain multiple steps together into a single object.
Here, our pipeline has two steps:
Scaler (
StandardScaler): ensures each feature has zero mean and unit variance, which helps many models train more effectively.Logistic Regression (
LogisticRegression): the actual linear classifier that fits a straight decision boundary to the data.
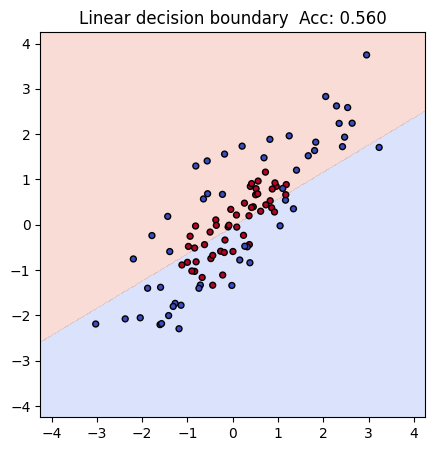
In our toy dataset, both x and y coordinates are on a similar numeric scale (roughly between -1.5 and 1.5). That means the results will look similar with or without the scaler. Still, you can see the first example has a higher acc 0.56 than the second one 0.54, and including it is good practice, because real-world datasets often have features on very different scales.
Xtr, Xte, ytr, yte = train_test_split(X_toy, y_toy, test_size=0.25, random_state=42, stratify=y_toy)
lin = Pipeline([
("scaler", StandardScaler()),
("logreg", LogisticRegression(max_iter=1000))
])
lin.fit(Xtr, ytr)
acc_lin = accuracy_score(yte, lin.predict(Xte))
print(f"Linear baseline accuracy with sacler: {acc_lin:.3f}")
Linear baseline accuracy with sacler: 0.560
By wrapping these steps in a pipeline, we can train, evaluate, and later reuse the combined process as if it were one model.
Without scaler, the result will be different in this dataset:
Xtr, Xte, ytr, yte = train_test_split(X_toy, y_toy, test_size=0.25, random_state=42, stratify=y_toy)
lin2 = LogisticRegression(max_iter=1000)
lin2.fit(Xtr, ytr)
acc_lin2 = accuracy_score(yte, lin2.predict(Xte))
print(f"Linear baseline accuracy without scaler: {acc_lin2:.3f}")
Linear baseline accuracy without scaler: 0.540
Regardless, as you can see, currently the prediction is pretty bad because linear regression itself does not handle this type of complex “donut” style dataset well due to its inherent limitation.
# Compact decision-boundary plot using only the first two features
# square limits from first two columns
lim = np.abs(np.vstack([Xtr[:, :2], Xte[:, :2]])).max()
pad = 0.5
xs = np.linspace(-lim - pad, lim + pad, 300)
ys = np.linspace(-lim - pad, lim + pad, 300)
xx, yy = np.meshgrid(xs, ys)
# build full grid (fill extra features with training means)
grid = np.c_[xx.ravel(), yy.ravel()]
if Xtr.shape[1] > 2:
rest = np.tile(Xtr[:, 2:].mean(axis=0), (grid.shape[0], 1))
grid = np.hstack([grid, rest])
Z = lin.predict(grid).reshape(xx.shape)
plt.figure(figsize=(5,5))
plt.contourf(xx, yy, Z, levels=2, alpha=0.25, cmap="coolwarm")
plt.scatter(Xte[:, 0], Xte[:, 1], c=yte, cmap="coolwarm", s=18, edgecolor="k")
plt.xlim(-lim - pad, lim + pad)
plt.ylim(-lim - pad, lim + pad)
plt.gca().set_aspect("equal", adjustable="box")
plt.title(f"Linear decision boundary Acc: {acc_lin:.3f}")
plt.show()
1.3 Build a small MLP step by step#
Now let’s see if we can solve this classification challenge with simple and shallow neural network.
We will build a MLP classifier by adding one hidden layer with 8 units and ReLU activation. Before we start to do it, we will first look at key concepts:
A multilayer perceptron (MLP) stacks simple building blocks:
Linear layer: multiply by weights and add a bias. Example:
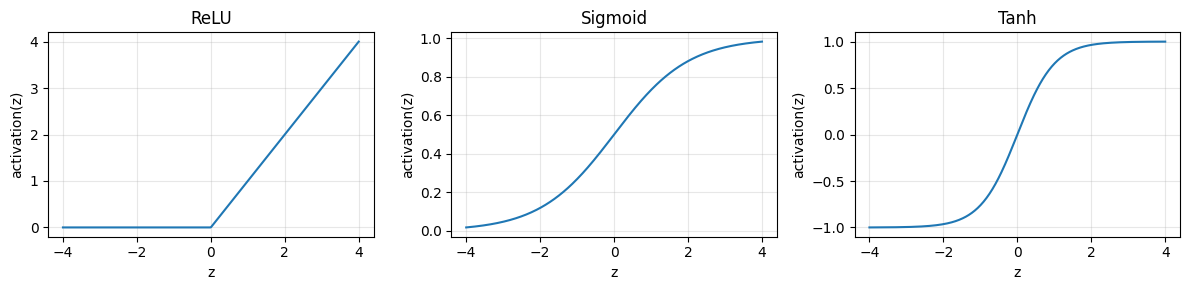
z = X @ W + bActivation: bend the space so a straight line can become a curve. Common choices:
ReLU:
max(0, z)Tanh: squashes to
[-1, 1]Sigmoid: squashes to
[0, 1]and is often used at the output for probability
Loss: tells us how wrong we are. For classification we use cross entropy
Optimizer: updates weights to reduce loss. Scikit-learn’s MLP uses SGD variants under the hood
Why ReLU
It is simple, fast, and avoids the flat gradients that slow learning with sigmoid on hidden layers
Many ReLU units stacked together can carve a curved boundary that a linear model cannot
Below we first look at the shapes of common activations, then we build a small MLP and read its learned shapes.
To keep it simple, imagine we have just one input \(x\), two hidden ReLU units, and one sigmoid output.
Note
Steps:
Hidden pre-activations
$\( z^{(1)}_1 = w^{(1)}_{1}\,x + b^{(1)}_{1}, \quad z^{(1)}_2 = w^{(1)}_{2}\,x + b^{(1)}_{2} \)$Hidden activations (apply ReLU)
$\( h_1 = \max(0, z^{(1)}_1), \quad h_2 = \max(0, z^{(1)}_2) \)$Output score and probability
$\( z^{(2)} = v_{1} h_1 + v_{2} h_2 + b^{(2)}, \quad \hat{p} = \sigma(z^{(2)}) = \frac{1}{1 + e^{-z^{(2)}}} \)$
During training, the model learns weights \(w^{(1)}_{1}, w^{(1)}_{2}, v_1, v_2\) and biases \(b^{(1)}_{1}, b^{(1)}_{2}, b^{(2)}\) to minimize classification error.
Let’s look at one example with numbers so it’s easier to understand:
Suppose:
Input: \(x = 0.5\)
Hidden weights and biases:
\(w^{(1)}_{1} = 2.0,\; b^{(1)}_{1} = -0.6\)
\(w^{(1)}_{2} = -1.0,\; b^{(1)}_{2} = 0.2\)Output weights and bias:
\(v_1 = 1.0,\; v_2 = -2.0,\; b^{(2)} = 0.1\)
Step 1. Hidden pre-activations
$\(
z^{(1)}_1 = 2.0 \cdot 0.5 + (-0.6) = 0.4
\)\(
\)\(
z^{(1)}_2 = -1.0 \cdot 0.5 + 0.2 = -0.3
\)$
Step 2. Hidden activations (ReLU)
$\(
h_1 = \max(0, 0.4) = 0.4
\)\(
\)\(
h_2 = \max(0, -0.3) = 0
\)$
Step 3. Output
$\(
z^{(2)} = 1.0 \cdot 0.4 + (-2.0) \cdot 0 + 0.1 = 0.5
\)\(
\)\(
\hat{p} = \sigma(0.5) = \frac{1}{1+e^{-0.5}} \approx 0.62
\)$
So with this set of weights, input \(x=0.5\) is classified with probability \(\hat{p} \approx 0.62\) for class 1.
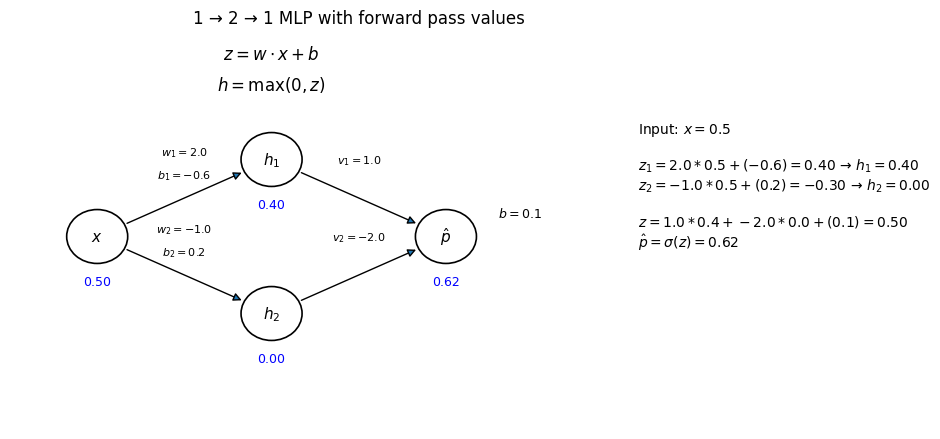
Note that in above example we already have a set of paramters so the calculation can be demonstrated.
In a real neural network, the weights \(w\) and biases \(b\) are not chosen by hand.
So, where do these numbers come from?
Try to recall what happens in linear regression.
In that case, you can compute the exact formulas for \(w\) and \(b\) by hand using sums and algebra or use lin.fit(Xtr, ytr) to get your trained function.
In a neural network, you usually don’t solve directly with formulas. Instead, you:
Initialize \(w\) and \(b\) randomly.
Adjust them step by step so that the loss (for example, mean squared error or cross-entropy) becomes smaller.
Run the next iteration with the updated weights, check the loss again, and adjust \(w\) and \(b\) further to aim for an even smaller loss.
After many updates, the parameters \(w\) and \(b\) converge to useful values — often close to what you would get from regression formulas in simple cases.
Because this process is iterative, the real learned numbers can look irregular, like \(w=0.483\) or \(b=-1.273\).
For teaching, we instead pick round numbers so it’s easier to see the flow of calculations by hand.
mlp = Pipeline([
("scaler", StandardScaler()),
("clf", MLPClassifier(hidden_layer_sizes=(8,),
activation="relu",
learning_rate_init=0.05,
alpha=1e-4, # L2 penalty
max_iter=500,
random_state=0))
])
mlp
Pipeline(steps=[('scaler', StandardScaler()),
('clf',
MLPClassifier(hidden_layer_sizes=(8,), learning_rate_init=0.05,
max_iter=500, random_state=0))])In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
Parameters
| steps | [('scaler', ...), ('clf', ...)] | |
| transform_input | None | |
| memory | None | |
| verbose | False |
Parameters
| copy | True | |
| with_mean | True | |
| with_std | True |
Parameters
| hidden_layer_sizes | (8,) | |
| activation | 'relu' | |
| solver | 'adam' | |
| alpha | 0.0001 | |
| batch_size | 'auto' | |
| learning_rate | 'constant' | |
| learning_rate_init | 0.05 | |
| power_t | 0.5 | |
| max_iter | 500 | |
| shuffle | True | |
| random_state | 0 | |
| tol | 0.0001 | |
| verbose | False | |
| warm_start | False | |
| momentum | 0.9 | |
| nesterovs_momentum | True | |
| early_stopping | False | |
| validation_fraction | 0.1 | |
| beta_1 | 0.9 | |
| beta_2 | 0.999 | |
| epsilon | 1e-08 | |
| n_iter_no_change | 10 | |
| max_fun | 15000 |
Fit and evaluate.
mlp.fit(Xtr, ytr)
acc_mlp = accuracy_score(yte, mlp.predict(Xte))
print(f"MLP accuracy: {acc_mlp:.3f}")
MLP accuracy: 0.890
Compared with linear regression (baseline accuracy: 0.560), we now get an MLP accuracy: 0.870, which is much higher!
1.5 Analyzing the MLP models#
Now let’s look at the plot below to see the structure of the MLP model we built.
Remember, we have 5 input x features. In the earlier plot we only showed the first two (X–Y) for visualization, but each point actually comes from all 5 features — that 2D scatter is just a projection. Think back to the earlier lecture where a molecule could be represented by multiple features such as MolWt, LogP, etc. In the same way, here all 5 features are connected to the hidden layer (with 5 units in this example).
Number of layers (including input and output): 3
Hidden layer sizes: (8,)
coefs_ length (weight matrices between layers): 2
Layer 1 weight shape: (5, 8) bias shape: (8,)
Layer 2 weight shape: (8, 1) bias shape: (1,)
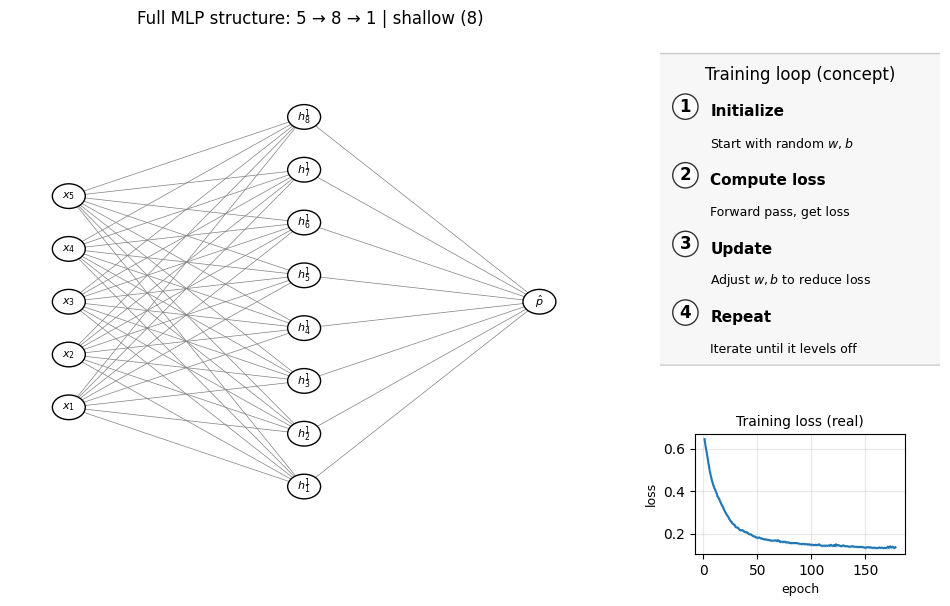
Does it mean that the more complex the MLP, the better results we can? Well, not necessaily. Look at the examlple below, where instead of just 1 layer we have 3 hidden layers (4 layers in total including the final output layer):
mlp_deep = Pipeline([
("scaler", StandardScaler()),
("clf", MLPClassifier(hidden_layer_sizes=(8,5,3),
activation="relu",
learning_rate_init=0.05,
max_iter=500,
random_state=0))
])
mlp_deep.fit(Xtr, ytr)
acc_mlp_deep = accuracy_score(yte, mlp_deep.predict(Xte))
print(f"MLP_deep accuracy: {acc_mlp_deep:.3f}")
summarize_mlp(mlp_deep); draw_full_mlp_structure(mlp_deep, Xtr, "deep (8,5,3)")
MLP_deep accuracy: 0.760
Number of layers (including input and output): 5
Hidden layer sizes: (8, 5, 3)
coefs_ length (weight matrices between layers): 4
Layer 1 weight shape: (5, 8) bias shape: (8,)
Layer 2 weight shape: (8, 5) bias shape: (5,)
Layer 3 weight shape: (5, 3) bias shape: (3,)
Layer 4 weight shape: (3, 1) bias shape: (1,)
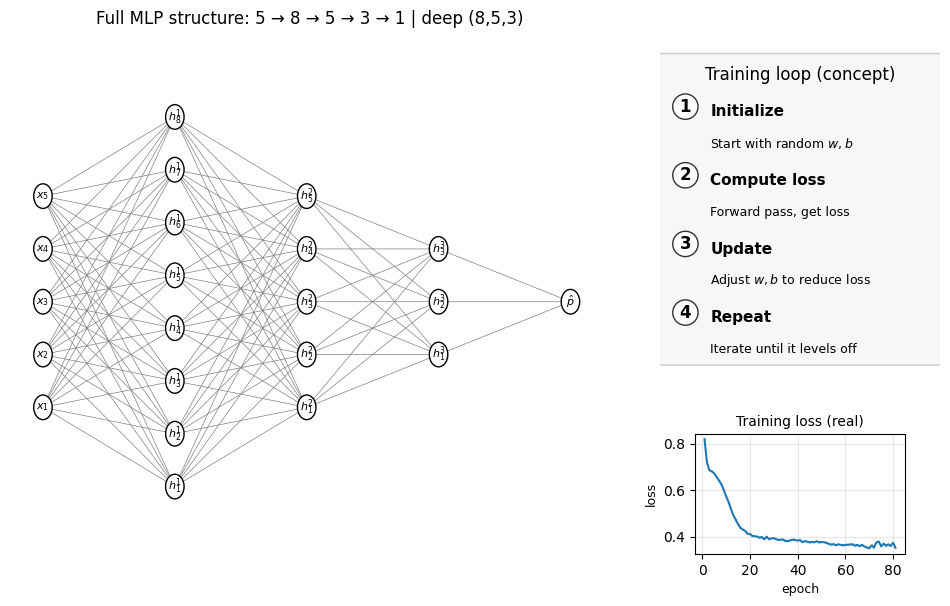
The accuray actually drops down! If you look at the loss function above, it’s also worse than the simple 5-5-1 shallow MLP we showed as the first example. Just recall the decision tree lecture we had before, the lesson is that you can’t just make super complex model to increase the accuracy because the overfitting issue will come at a certain point.
But it doesn’t necessarily mean we shouldn’t do it. Below is another case and we actually achieve a higher performance.
mlp_deep_2 = Pipeline([
("scaler", StandardScaler()),
("clf", MLPClassifier(hidden_layer_sizes=(8,4,6,4),
activation="relu",
learning_rate_init=0.05,
max_iter=500,
random_state=0))
])
mlp_deep_2.fit(Xtr, ytr)
acc_mlp_deep_2 = accuracy_score(yte, mlp_deep_2.predict(Xte))
print(f"MLP_deep_2 accuracy: {acc_mlp_deep_2:.3f}")
summarize_mlp(mlp_deep_2); draw_full_mlp_structure(mlp_deep_2, Xtr, "deep (8,4,6,4)")
MLP_deep_2 accuracy: 0.870
Number of layers (including input and output): 6
Hidden layer sizes: (8, 4, 6, 4)
coefs_ length (weight matrices between layers): 5
Layer 1 weight shape: (5, 8) bias shape: (8,)
Layer 2 weight shape: (8, 4) bias shape: (4,)
Layer 3 weight shape: (4, 6) bias shape: (6,)
Layer 4 weight shape: (6, 4) bias shape: (4,)
Layer 5 weight shape: (4, 1) bias shape: (1,)
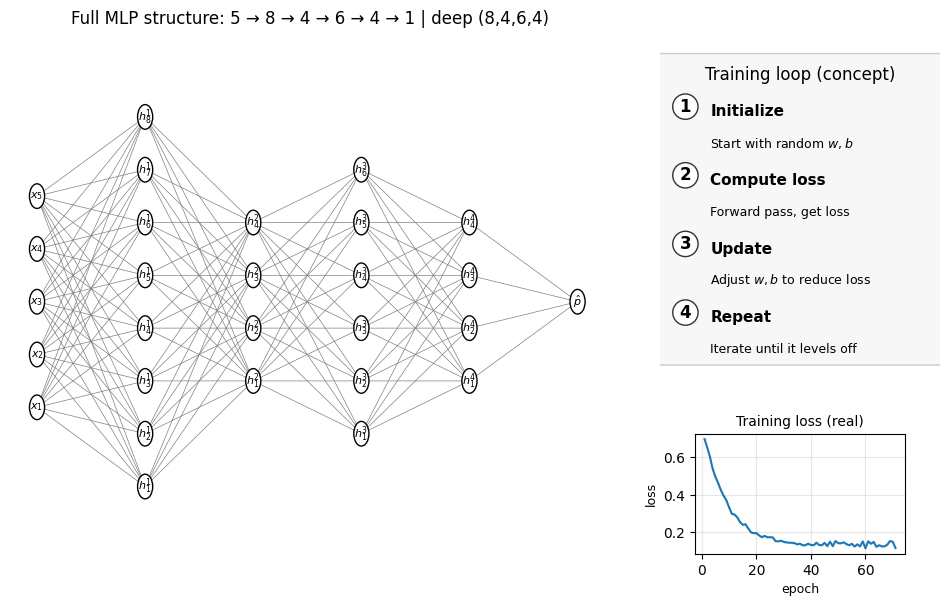
1.6 Decision surface#
We can plot a decision surface to see the boundary and compare all these three models.
import numpy as np
import matplotlib.pyplot as plt
# square limits from the first two columns
lim = np.abs(np.vstack([Xtr[:, :2], Xte[:, :2]])).max()
pad = 0.5
xs = np.linspace(-lim - pad, lim + pad, 300)
ys = np.linspace(-lim - pad, lim + pad, 300)
xx, yy = np.meshgrid(xs, ys)
# 2D grid for the first two features
grid2 = np.c_[xx.ravel(), yy.ravel()]
# fill the remaining features with training means (raw space)
if Xtr.shape[1] > 2:
rest_means = Xtr[:, 2:].mean(axis=0) # shape (3,)
rest = np.tile(rest_means, (grid2.shape[0], 1)) # shape (N, 3)
grid_full = np.hstack([grid2, rest]) # shape (N, 5)
else:
grid_full = grid2
# probabilities for both models
zz_shallow = mlp.predict_proba(grid_full)[:, 1].reshape(xx.shape)
zz_deep = mlp_deep.predict_proba(grid_full)[:, 1].reshape(xx.shape)
zz_deep_2 = mlp_deep_2.predict_proba(grid_full)[:, 1].reshape(xx.shape)
# plot side by side
fig, axes = plt.subplots(1, 3, figsize=(12,5), sharex=True, sharey=True)
for ax, zz, title in zip(
axes,
[zz_shallow, zz_deep, zz_deep_2],
["Shallow MLP (5 hidden)", "Deep MLP (8,5,3 hidden)", "Deep MLP (8, 4, 6, 4)"]
):
cs = ax.contourf(xx, yy, zz, levels=20, alpha=0.4, cmap="coolwarm") # 50% transparency
ax.scatter(Xtr[:,0], Xtr[:,1], c=ytr, cmap="coolwarm", s=3, alpha=0.7, label="train")
ax.scatter(Xte[:,0], Xte[:,1], c=yte, cmap="coolwarm", s=3, marker="^", label="test")
ax.set_aspect("equal", adjustable="box")
ax.set_title(title)
fig.colorbar(cs, ax=axes, label="P(class=1)")
axes[0].legend(loc="lower right")
plt.show()
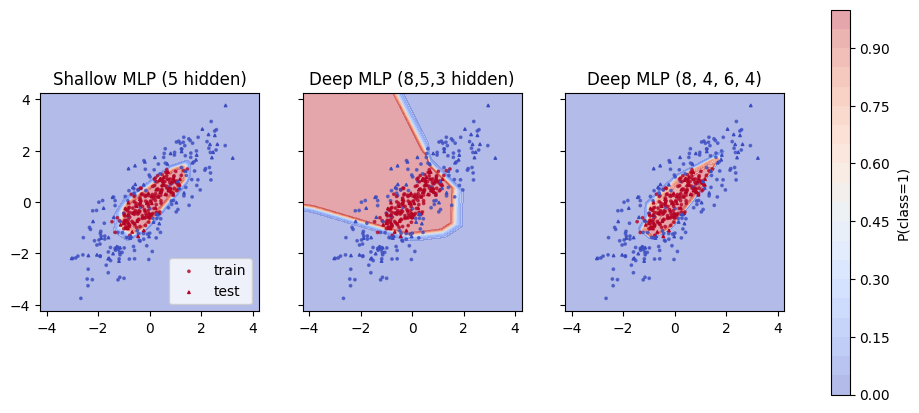
Color near 0 is class 0 and near 1 is class 1. A curved boundary shows that the hidden layer and activation created a nonlinear rule.
⏰ Exercises 1
Change
hidden_layer_sizes=(16,)then(16,8)and compare test accuracy.Switch
activationfrom"relu"to"tanh"and rerun. Which works better here and why might that be?Change
learning_rate_initto0.005then0.2. Watch whether the model converges. If it stalls, raisemax_iter.Remove the scaler from the pipeline and fit again. Record the accuracy. What changed?
1.7 Hyperparamter tuning#
As we see in previous lectures, we can use cross-validation to search for the best hyperparamters. In the MLP example below, to demonstrate this idea, we can try different layer choice and learning rate. But keep in mind that there are multiple hyperparameters we can also tune. To make the demo simple we just fix most of them.
Below code will take around 2 minutes to complete, since it’s alreay 10 x 4 = 40 choices of hyperparamters.
Fitting 5 folds for each of 80 candidates, totalling 400 fits
Best CV accuracy: 0.890
Best params: {'clf__hidden_layer_sizes': (np.int64(16), 16), 'clf__learning_rate_init': 0.001}
Train accuracy (refit best): 0.953
Test accuracy (refit best): 0.890
| hidden_layer_sizes | learning_rate_init | mean_test_score | std_test_score | rank_test_score | |
|---|---|---|---|---|---|
| 0 | (16, 16) | 0.001 | 0.890000 | 0.027080 | 1 |
| 1 | (8, 6, 3) | 0.050 | 0.886667 | 0.038586 | 2 |
| 2 | (24, 4, 4, 4, 4) | 0.020 | 0.880000 | 0.012472 | 3 |
| 3 | (16, 16) | 0.050 | 0.876667 | 0.034319 | 4 |
| 4 | (32, 24) | 0.001 | 0.876667 | 0.037417 | 4 |
| 5 | (12, 8) | 0.010 | 0.876667 | 0.030912 | 4 |
| 6 | (32, 28) | 0.001 | 0.873333 | 0.038873 | 7 |
| 7 | (8, 6, 3) | 0.010 | 0.873333 | 0.024944 | 7 |
| 8 | (8, 6, 3) | 0.020 | 0.870000 | 0.019437 | 9 |
| 9 | (16, 8, 4) | 0.001 | 0.866667 | 0.027889 | 10 |
Number of layers (including input and output): 4
Hidden layer sizes: (np.int64(16), 16)
coefs_ length (weight matrices between layers): 3
Layer 1 weight shape: (5, 16) bias shape: (16,)
Layer 2 weight shape: (16, 16) bias shape: (16,)
Layer 3 weight shape: (16, 1) bias shape: (1,)
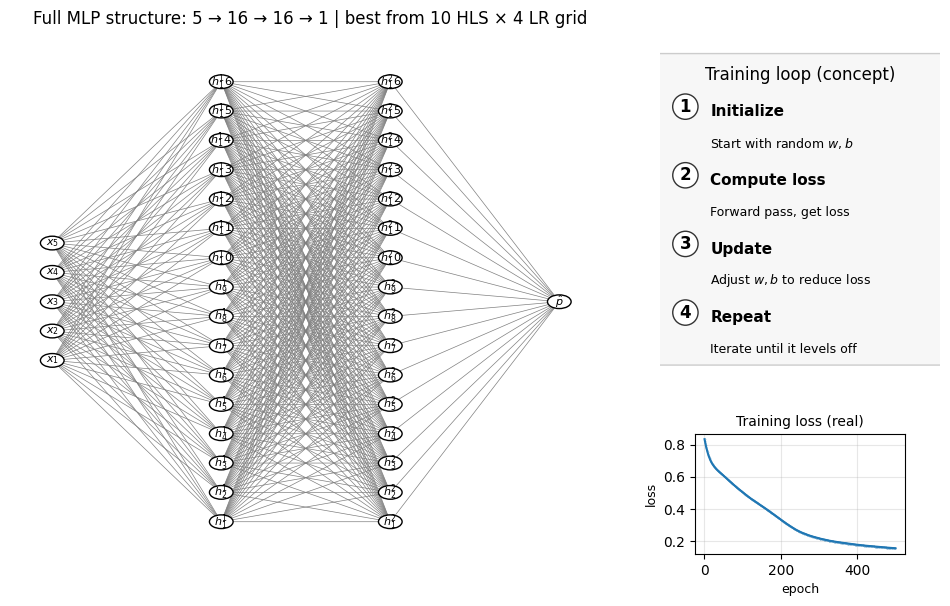
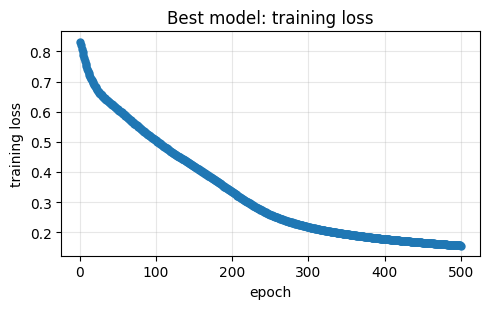
2. Neural nets for chemical property prediction#
We reuse the same CSV from earlier lectures and compute four quick descriptors.
url = "https://raw.githubusercontent.com/zzhenglab/ai4chem/main/book/_data/C_H_oxidation_dataset.csv"
df_raw = pd.read_csv(url)
df_raw.head(3)
| Compound Name | CAS | SMILES | Solubility_mol_per_L | pKa | Toxicity | Melting Point | Reactivity | Oxidation Site | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 3,4-dihydro-1H-isochromene | 493-05-0 | c1ccc2c(c1)CCOC2 | 0.103906 | 5.80 | non_toxic | 65.8 | 1 | 8,10 |
| 1 | 9H-fluorene | 86-73-7 | c1ccc2c(c1)Cc1ccccc1-2 | 0.010460 | 5.82 | toxic | 90.0 | 1 | 7 |
| 2 | 1,2,3,4-tetrahydronaphthalene | 119-64-2 | c1ccc2c(c1)CCCC2 | 0.020589 | 5.74 | toxic | 69.4 | 1 | 7,10 |
def calc_desc(smiles):
if Chem is None:
return pd.Series({"MolWt": np.nan, "LogP": np.nan, "TPSA": np.nan, "NumRings": np.nan})
m = Chem.MolFromSmiles(smiles)
if m is None:
return pd.Series({"MolWt": np.nan, "LogP": np.nan, "TPSA": np.nan, "NumRings": np.nan})
return pd.Series({
"MolWt": Descriptors.MolWt(m),
"LogP": Crippen.MolLogP(m),
"TPSA": rdMolDescriptors.CalcTPSA(m),
"NumRings": rdMolDescriptors.CalcNumRings(m)
})
desc = df_raw["SMILES"].apply(calc_desc)
df = pd.concat([df_raw, desc], axis=1)
df[["Compound Name","SMILES","MolWt","LogP","TPSA","NumRings","Melting Point","Solubility_mol_per_L","Toxicity"]].head()
| Compound Name | SMILES | MolWt | LogP | TPSA | NumRings | Melting Point | Solubility_mol_per_L | Toxicity | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 3,4-dihydro-1H-isochromene | c1ccc2c(c1)CCOC2 | 134.178 | 1.7593 | 9.23 | 2.0 | 65.8 | 0.103906 | non_toxic |
| 1 | 9H-fluorene | c1ccc2c(c1)Cc1ccccc1-2 | 166.223 | 3.2578 | 0.00 | 3.0 | 90.0 | 0.010460 | toxic |
| 2 | 1,2,3,4-tetrahydronaphthalene | c1ccc2c(c1)CCCC2 | 132.206 | 2.5654 | 0.00 | 2.0 | 69.4 | 0.020589 | toxic |
| 3 | ethylbenzene | CCc1ccccc1 | 106.168 | 2.2490 | 0.00 | 1.0 | 65.0 | 0.048107 | non_toxic |
| 4 | cyclohexene | C1=CCCCC1 | 82.146 | 2.1166 | 0.00 | 1.0 | 96.4 | 0.060688 | non_toxic |
Reminder
We used these same descriptors in Lectures 5 to 7. They form a compact input vector for both regression and classification.
Neural nets are sensitive to feature scales. Always standardize inputs.
We will show MLP performance with and without StandardScaler on a quick regression target.
2.1 Target choice: melting point#
df_reg = df[["MolWt","LogP","TPSA","NumRings","Melting Point"]].dropna()
X = df_reg[["MolWt","LogP","TPSA","NumRings"]].values
y = df_reg["Melting Point"].values
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
X_train.shape, X_test.shape
((460, 4), (115, 4))
MLP without scaling:
mlp_raw = MLPRegressor(hidden_layer_sizes=(64,32),
activation="relu",
learning_rate_init=0.01,
alpha=1e-4,
max_iter=1000,
random_state=0)
mlp_raw.fit(X_train, y_train)
y_pred_raw = mlp_raw.predict(X_test)
print(f"R2 no scaling: {r2_score(y_test, y_pred_raw):.3f}")
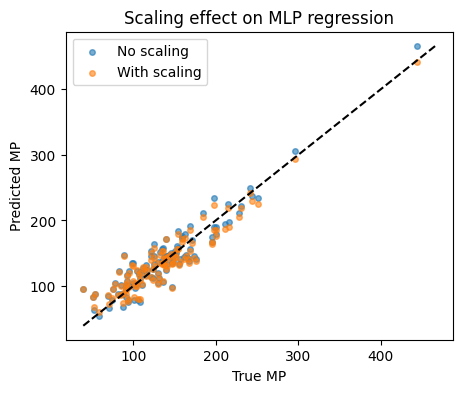
R2 no scaling: 0.867
MLP with scaling:
mlp_scaled = Pipeline([
("scaler", StandardScaler()),
("reg", MLPRegressor(hidden_layer_sizes=(64,32),
activation="relu",
learning_rate_init=0.01,
alpha=1e-4,
max_iter=1000,
random_state=0))
])
mlp_scaled.fit(X_train, y_train)
y_pred_scaled = mlp_scaled.predict(X_test)
print(f"R2 with scaling: {r2_score(y_test, y_pred_scaled):.3f}")
R2 with scaling: 0.873
Parity plots to compare:
⏰ Exercises 2
Try a single hidden layer
(32,)and two layers(128,64). Record R². Which is better here?Change
alphato1e-3then1e-2. This is L2 regularization. How does it change test R² and the gap between train and test?Set
early_stopping=Trueandvalidation_fraction=0.15. Compare training time and R².
3. Regression with MLP on log-solubility#
Solubility has a long tail. We saw in Lecture 5 that log transform helped.
3.1 Prepare data#
df_sol = df.copy()
df_sol = df_sol[["MolWt","LogP","TPSA","NumRings","Solubility_mol_per_L"]].dropna()
df_sol["logS"] = np.log10(df_sol["Solubility_mol_per_L"] + 1e-6)
X = df_sol[["MolWt","LogP","TPSA","NumRings"]].values
y = df_sol["logS"].values
Xtr, Xte, ytr, yte = train_test_split(X, y, test_size=0.2, random_state=15)
Xtr[:3], ytr[:5]
(array([[174.243 , 2.768 , 17.07 , 2. ],
[250.725 , 3.6854, 26.3 , 3. ],
[316.154 , 3.2662, 46.17 , 3. ]]),
array([-1.33079381, -1.81944953, -1.66573637, -1.73461148, -1.83886568]))
3.2 Build the pipeline and fit#
We start small then expand.
reg1 = Pipeline([
("scaler", StandardScaler()),
("mlp", MLPRegressor(hidden_layer_sizes=(32,),
activation="relu",
learning_rate_init=0.01,
alpha=1e-3,
max_iter=1000,
random_state=0))
])
reg1
Pipeline(steps=[('scaler', StandardScaler()),
('mlp',
MLPRegressor(alpha=0.001, hidden_layer_sizes=(32,),
learning_rate_init=0.01, max_iter=1000,
random_state=0))])In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
Parameters
| steps | [('scaler', ...), ('mlp', ...)] | |
| transform_input | None | |
| memory | None | |
| verbose | False |
Parameters
| copy | True | |
| with_mean | True | |
| with_std | True |
Parameters
| loss | 'squared_error' | |
| hidden_layer_sizes | (32,) | |
| activation | 'relu' | |
| solver | 'adam' | |
| alpha | 0.001 | |
| batch_size | 'auto' | |
| learning_rate | 'constant' | |
| learning_rate_init | 0.01 | |
| power_t | 0.5 | |
| max_iter | 1000 | |
| shuffle | True | |
| random_state | 0 | |
| tol | 0.0001 | |
| verbose | False | |
| warm_start | False | |
| momentum | 0.9 | |
| nesterovs_momentum | True | |
| early_stopping | False | |
| validation_fraction | 0.1 | |
| beta_1 | 0.9 | |
| beta_2 | 0.999 | |
| epsilon | 1e-08 | |
| n_iter_no_change | 10 | |
| max_fun | 15000 |
Note: You can click on MLPRehressor above to see the details.
3.3 Parity and training curve#
MLPRegressor exposes loss_curve_ after fit. This is the average loss per epoch on the training data.
Compare with lasso (optimized with alpha = 0.1) we learned in Lecture 5, you can see its performance is better even without optimizing the hyperparameters.
reg1.fit(Xtr, ytr)
yhat = reg1.predict(Xte)
print(f"MSE: {mean_squared_error(yte, yhat):.4f}")
print(f"MAE: {mean_absolute_error(yte, yhat):.4f}")
print(f"R2: {r2_score(yte, yhat):.3f}")
MSE: 0.0185
MAE: 0.1075
R2: 0.966
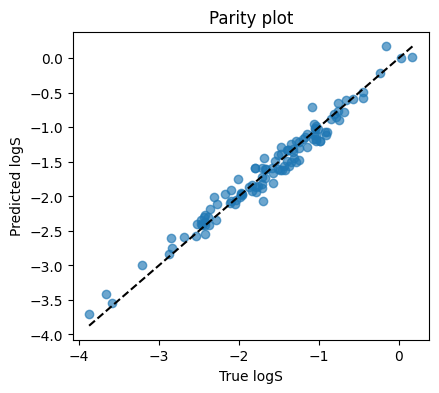
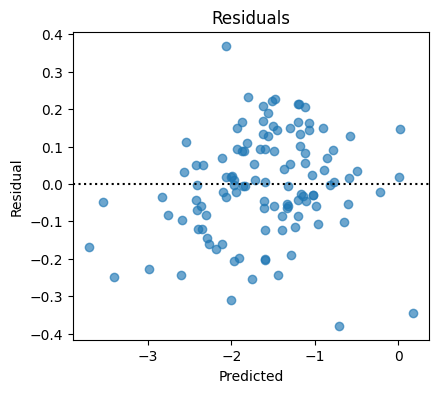
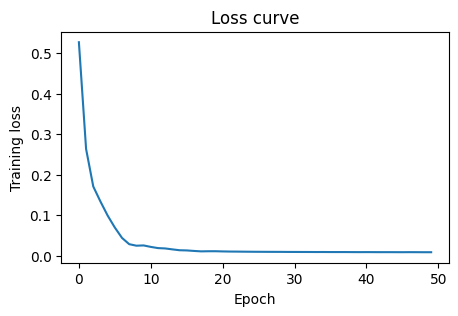
4. Classification with MLP on toxicity#
Moving beyond regression, let’s look an example on the classification.
We will predict toxic vs non_toxic from the same four descriptors.
4.1 Prepare labels and build classifier#
df_clf = df[["MolWt","LogP","TPSA","NumRings","Toxicity"]].dropna()
label_map = {"toxic":1, "non_toxic":0}
y = df_clf["Toxicity"].str.lower().map(label_map).astype(int).values
X = df_clf[["MolWt","LogP","TPSA","NumRings"]].values
Xtr, Xte, ytr, yte = train_test_split(X, y, test_size=0.2, random_state=42, stratify=y)
clf_pipe = Pipeline([
("scaler", StandardScaler()),
("mlp", MLPClassifier(hidden_layer_sizes=(16,),
activation="relu",
learning_rate_init=0.01,
max_iter=2000,
random_state=0))
])
clf_pipe
Pipeline(steps=[('scaler', StandardScaler()),
('mlp',
MLPClassifier(hidden_layer_sizes=(16,),
learning_rate_init=0.01, max_iter=2000,
random_state=0))])In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
Parameters
| steps | [('scaler', ...), ('mlp', ...)] | |
| transform_input | None | |
| memory | None | |
| verbose | False |
Parameters
| copy | True | |
| with_mean | True | |
| with_std | True |
Parameters
| hidden_layer_sizes | (16,) | |
| activation | 'relu' | |
| solver | 'adam' | |
| alpha | 0.0001 | |
| batch_size | 'auto' | |
| learning_rate | 'constant' | |
| learning_rate_init | 0.01 | |
| power_t | 0.5 | |
| max_iter | 2000 | |
| shuffle | True | |
| random_state | 0 | |
| tol | 0.0001 | |
| verbose | False | |
| warm_start | False | |
| momentum | 0.9 | |
| nesterovs_momentum | True | |
| early_stopping | False | |
| validation_fraction | 0.1 | |
| beta_1 | 0.9 | |
| beta_2 | 0.999 | |
| epsilon | 1e-08 | |
| n_iter_no_change | 10 | |
| max_fun | 15000 |
Everything stays the same except we use MLPClassifier() instead of MLPRegressor()
Now let’s fit and evaluate.
clf_pipe.fit(Xtr, ytr)
yp = clf_pipe.predict(Xte)
yproba = clf_pipe.predict_proba(Xte)[:,1]
acc = accuracy_score(yte, yp)
prec = precision_score(yte, yp)
rec = recall_score(yte, yp)
f1 = f1_score(yte, yp)
auc = roc_auc_score(yte, yproba)
print(f"Accuracy: {acc:.3f}")
print(f"Precision: {prec:.3f}")
print(f"Recall: {rec:.3f}")
print(f"F1: {f1:.3f}")
print(f"AUC: {auc:.3f}")
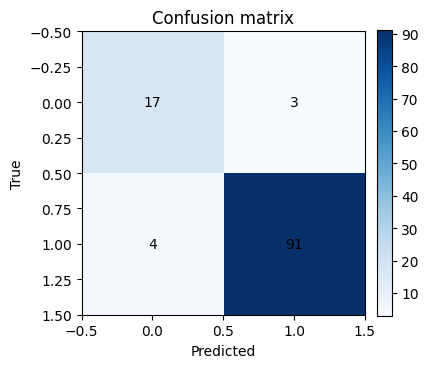
cm = confusion_matrix(yte, yp)
plt.figure(figsize=(4,4))
plt.imshow(cm, cmap="Blues")
plt.title("Confusion matrix")
plt.xlabel("Predicted")
plt.ylabel("True")
for i in range(2):
for j in range(2):
plt.text(j, i, cm[i,j], ha="center", va="center")
plt.colorbar(fraction=0.046, pad=0.04)
plt.show()
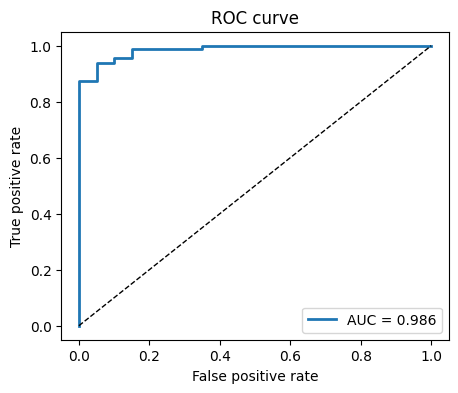
fpr, tpr, thr = roc_curve(yte, yproba)
plt.figure(figsize=(5,4))
plt.plot(fpr, tpr, lw=2, label=f"AUC = {auc:.3f}")
plt.plot([0,1], [0,1], "k--", lw=1)
plt.xlabel("False positive rate")
plt.ylabel("True positive rate")
plt.title("ROC curve")
plt.legend()
plt.show()
Accuracy: 0.939
Precision: 0.968
Recall: 0.958
F1: 0.963
AUC: 0.986
Threshold tuning demo.
for t in [0.3, 0.5, 0.7]:
yp_t = (yproba >= t).astype(int)
print(f"threshold={t:.2f} acc={accuracy_score(yte, yp_t):.3f} prec={precision_score(yte, yp_t):.3f} rec={recall_score(yte, yp_t):.3f}")
threshold=0.30 acc=0.965 prec=0.969 rec=0.989
threshold=0.50 acc=0.939 prec=0.968 rec=0.958
threshold=0.70 acc=0.930 prec=0.978 rec=0.937
5. Peek inside the network#
While scikit-learn MLP is a black box in training, you can still inspect weights.
We will look at the first layer weights for regression on log-solubility and see which descriptors each hidden unit is sensitive to.
reg_inspect = Pipeline([
("scaler", StandardScaler()),
("mlp", MLPRegressor(hidden_layer_sizes=(8,),
activation="tanh",
learning_rate_init=0.01,
alpha=1e-3,
max_iter=1500,
random_state=0))
]).fit(Xtr, ytr)
W1 = reg_inspect.named_steps["mlp"].coefs_[0] # shape (4, 8)
cols = ["MolWt","LogP","TPSA","NumRings"]
plt.figure(figsize=(6,3.5))
plt.imshow(W1, aspect="auto", cmap="bwr")
plt.yticks(range(len(cols)), cols)
plt.xticks(range(W1.shape[1]), [f"h{j}" for j in range(W1.shape[1])])
plt.colorbar(label="weight")
plt.title("First layer weights")
plt.show()
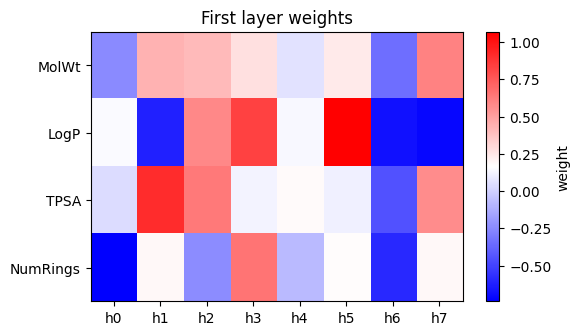
How to read above plot:
Each column is a hidden unit. Positive weights mean the unit activates when that feature is larger than average. Negative weights mean the opposite. This is not a full explanation, but it gives a flavor of what the network latches onto.
⏰ Exercises 5
Try to overwrite the Xtr, ytr with different task (e.g. melting point regression, solubility regression, toxicity classification) and use different MLP to see the difference.
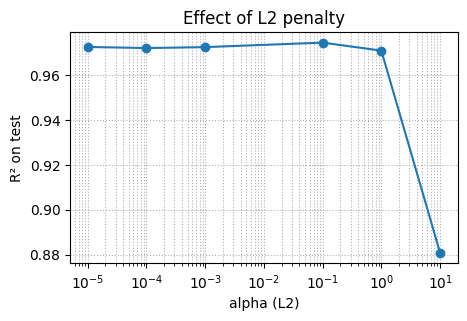
6. Overfitting, regularization, and early stopping#
Neural nets can overfit small chemistry tables. Two easy controls:
L2 penalty via
alpha. Larger alpha shrinks weights.Early stopping stops when validation score stops improving.
We will show a sweep of alpha for log-solubility.
7. Glossary#
- multilayer perceptron#
A network of layers that apply linear transforms plus nonlinear activations.
- activation#
Nonlinear function applied elementwise. Common choices: ReLU, tanh, logistic.
- loss#
A number that measures how wrong the predictions are on training data.
- optimizer#
Method that updates weights to reduce loss. Scikit-learn uses variants of SGD or LBFGS under the hood.
- alpha#
L2 penalty strength in scikit-learn MLP. Larger means more shrinkage.
- early stopping#
Stop training when validation score does not improve for a patience window.
- scaling#
Standardize features to zero mean and unit variance. Critical for neural nets.
- parity plot#
Scatter of predicted vs true values for regression.
- ROC AUC#
Area under the Receiver Operating Characteristic curve. Threshold free ranking metric.
A layer between input and output that lets the model learn nonlinear combinations.
8. Quick reference#
Recipes
Always scale inputs for MLPs:
Pipeline([("scaler", StandardScaler()), ("mlp", ...)])Start small: one hidden layer with 16 to 64 units
For regression: check parity and residual plots
For classification: look at confusion matrix and ROC
To avoid overfitting: increase
alphaand useearly_stopping=TrueReproducibility: set
random_state
Common args
hidden_layer_sizes=(h1, h2, ...)number of units per layeractivation="relu"or"tanh"alphaL2 penalty, try 1e-5 to 1e-2learning_rate_initstep size, try 0.001 to 0.05max_itercap on epochsearly_stopping=Trueandvalidation_fraction=0.1 to 0.2
9. In-class activity#
Q1. Tiny MLP for melting point#
Use features
[MolWt, LogP, TPSA, NumRings]Split 80/20 with
random_state=7Train
MLPRegressorwith(32,),alpha=1e-3, ReLUReport
MSE,MAE,R²and draw a parity plot
# Q1 starter
...
df_reg = df[["MolWt","LogP","TPSA","NumRings","Melting Point"]].dropna()
X = df_reg[["MolWt","LogP","TPSA","NumRings"]].values
y = df_reg["Melting Point"].values
Xtr, Xte, ytr, yte = train_test_split(...) #TO DO
pipe = Pipeline(...).fit(Xtr, ytr) #TO DO
yhat = pipe.predict(Xte)
print(f"MSE={mean_squared_error(yte,yhat):.2f} MAE={mean_absolute_error(yte,yhat):.2f} R2={r2_score(yte,yhat):.3f}")
plt.figure(figsize=(4.5,4))
plt.scatter(yte, yhat, alpha=0.65)
lims = [min(yte.min(), yhat.min()), max(yte.max(), yhat.max())]
plt.plot(lims, lims, "k--")
plt.xlabel("True MP"); plt.ylabel("Pred MP"); plt.title("Q1 parity")
plt.show()
Q2. Depth vs width on log-solubility#
Train three models on
logSwith hidden sizes(16,),(32,),(64,32)Keep
alpha=1e-3,learning_rate_init=0.01, early stopping onCompare test
R²and plot the three loss curves on the same plot
# Q2 starter
df_sol = df[["MolWt","LogP","TPSA","NumRings","Solubility_mol_per_L"]].dropna().copy()
df_sol["logS"] = np.log10(df_sol["Solubility_mol_per_L"]+1e-6)
X = df_sol[["MolWt","LogP","TPSA","NumRings"]].values
y = df_sol["logS"].values
Xtr, Xte, ytr, yte = train_test_split(X, y, test_size=0.2, random_state=15)
sizes = [...] #TO DO
r2s, curves = [], []
for sz in sizes:
pipe = Pipeline([
("scaler", StandardScaler()),
("mlp", MLPRegressor(hidden_layer_sizes=sz, activation="relu",
alpha=..., #TO DO
learning_rate_init=..., #TO DO
early_stopping=..., #TO DO
validation_fraction=0.15,
max_iter=3000, random_state=0))
]).fit(Xtr, ytr)
yhat = pipe.predict(Xte)
r2s.append(r2_score(yte, yhat))
curves.append(pipe.named_steps["mlp"].loss_curve_)
print(pd.DataFrame({"hidden_sizes":[str(s) for s in sizes],"R2":np.round(r2s,3)}))
plt.figure(figsize=(5.5,3.5))
for sz, c in zip(sizes, curves):
plt.plot(c, label=str(sz))
plt.xlabel("Epoch"); plt.ylabel("Loss"); plt.title("Q2 loss curves")
plt.legend(); plt.show()
Q3. Toxicity classification with threshold tuning#
For
Toxicity, trainMLPClassifierwith(32,),alpha=1e-3, early stopping onCompute probabilities on test
For thresholds
[0.3, 0.5, 0.7]print Accuracy, Precision, Recall, F1
#TO DO
Q4. Compare MLP to Linear Regression on logS#
Same train test split as Q2
Fit Linear Regression on scaled inputs for
logSReport both R² on test and draw both parity scatters on one plot
# TO DO
11. Solutions#
Solution Q1#
df_reg = df[["MolWt","LogP","TPSA","NumRings","Melting Point"]].dropna()
X = df_reg[["MolWt","LogP","TPSA","NumRings"]].values
y = df_reg["Melting Point"].values
Xtr, Xte, ytr, yte = train_test_split(X, y, test_size=0.2, random_state=7)
pipe = Pipeline([
("scaler", StandardScaler()),
("mlp", MLPRegressor(hidden_layer_sizes=(32,), activation="relu",
alpha=1e-3, learning_rate_init=0.01,
max_iter=1500, random_state=0))
]).fit(Xtr, ytr)
yhat = pipe.predict(Xte)
print(f"MSE={mean_squared_error(yte,yhat):.2f} MAE={mean_absolute_error(yte,yhat):.2f} R2={r2_score(yte,yhat):.3f}")
plt.figure(figsize=(4.5,4))
plt.scatter(yte, yhat, alpha=0.65)
lims = [min(yte.min(), yhat.min()), max(yte.max(), yhat.max())]
plt.plot(lims, lims, "k--")
plt.xlabel("True MP"); plt.ylabel("Pred MP"); plt.title("Q1 parity")
plt.show()
MSE=409.35 MAE=16.27 R2=0.855
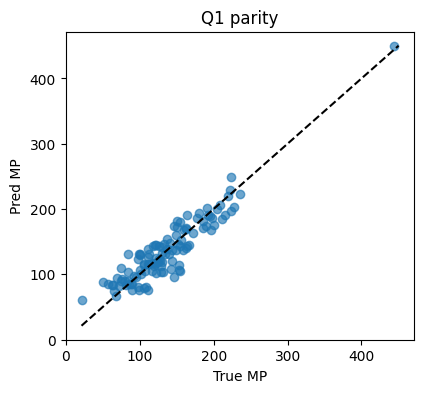
Solution Q2#
sizes = [(16,), (32,), (64,32)]
df_sol = df[["MolWt","LogP","TPSA","NumRings","Solubility_mol_per_L"]].dropna().copy()
df_sol["logS"] = np.log10(df_sol["Solubility_mol_per_L"]+1e-6)
X = df_sol[["MolWt","LogP","TPSA","NumRings"]].values
y = df_sol["logS"].values
Xtr, Xte, ytr, yte = train_test_split(X, y, test_size=0.2, random_state=15)
r2s, curves = [], []
for sz in sizes:
reg = Pipeline([
("scaler", StandardScaler()),
("mlp", MLPRegressor(hidden_layer_sizes=sz, activation="relu",
alpha=1e-3, learning_rate_init=0.01,
early_stopping=True, validation_fraction=0.15,
max_iter=3000, random_state=0))
]).fit(Xtr, ytr)
yhat = reg.predict(Xte)
r2s.append(r2_score(yte, yhat))
curves.append(reg.named_steps["mlp"].loss_curve_)
print(pd.DataFrame({"hidden_sizes":[str(s) for s in sizes],"R2":np.round(r2s,3)}))
plt.figure(figsize=(5.5,3.5))
for sz, c in zip(sizes, curves):
plt.plot(c, label=str(sz))
plt.xlabel("Epoch"); plt.ylabel("Loss"); plt.title("Q2 loss curves")
plt.legend(); plt.show()
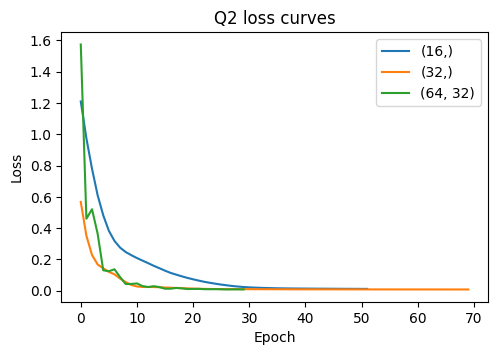
hidden_sizes R2
0 (16,) 0.950
1 (32,) 0.964
2 (64, 32) 0.958
Solution Q3#
df_clf = df[["MolWt","LogP","TPSA","NumRings","Toxicity"]].dropna()
y = df_clf["Toxicity"].str.lower().map({"toxic":1,"non_toxic":0}).astype(int).values
X = df_clf[["MolWt","LogP","TPSA","NumRings"]].values
Xtr, Xte, ytr, yte = train_test_split(X, y, test_size=0.2, random_state=42, stratify=y)
clf = Pipeline([
("scaler", StandardScaler()),
("mlp", MLPClassifier(hidden_layer_sizes=(32,), activation="relu",
alpha=1e-3, learning_rate_init=0.01,
early_stopping=True, validation_fraction=0.15,
max_iter=3000, random_state=0))
]).fit(Xtr, ytr)
proba = clf.predict_proba(Xte)[:,1]
for t in [0.3, 0.5, 0.7]:
pred = (proba >= t).astype(int)
print(f"t={t:.1f} acc={accuracy_score(yte,pred):.3f} prec={precision_score(yte,pred):.3f} rec={recall_score(yte,pred):.3f} f1={f1_score(yte,pred):.3f}")
t=0.3 acc=0.887 prec=0.887 rec=0.989 f1=0.935
t=0.5 acc=0.896 prec=0.946 rec=0.926 f1=0.936
t=0.7 acc=0.878 prec=0.976 rec=0.874 f1=0.922
Solution Q4#
df_sol = df[["MolWt","LogP","TPSA","NumRings","Solubility_mol_per_L"]].dropna().copy()
df_sol["logS"] = np.log10(df_sol["Solubility_mol_per_L"]+1e-6)
X = df_sol[["MolWt","LogP","TPSA","NumRings"]].values
y = df_sol["logS"].values
Xtr, Xte, ytr, yte = train_test_split(X, y, test_size=0.2, random_state=15)
sc = StandardScaler().fit(Xtr)
Xtr_s, Xte_s = sc.transform(Xtr), sc.transform(Xte)
lr = LinearRegression().fit(Xtr_s, ytr)
yhat_lr = lr.predict(Xte_s)
mlp = MLPRegressor(hidden_layer_sizes=(32,), activation="relu",
alpha=1e-3, learning_rate_init=0.01,
max_iter=3000, random_state=0).fit(Xtr_s, ytr)
yhat_mlp = mlp.predict(Xte_s)
print(f"Linear R2: {r2_score(yte, yhat_lr):.3f}")
print(f"MLP R2: {r2_score(yte, yhat_mlp):.3f}")
plt.figure(figsize=(5.5,4))
plt.scatter(yte, yhat_lr, alpha=0.6, label="Linear")
plt.scatter(yte, yhat_mlp, alpha=0.6, label="MLP")
lims = [min(yte.min(), yhat_lr.min(), yhat_mlp.min()), max(yte.max(), yhat_lr.max(), yhat_mlp.max())]
plt.plot(lims, lims, "k--")
plt.xlabel("True logS"); plt.ylabel("Predicted")
plt.legend(); plt.title("Q4 parity: Linear vs MLP")
plt.show()
Linear R2: 0.970
MLP R2: 0.966
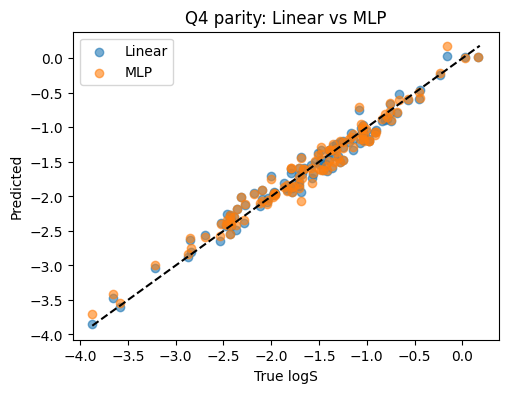
Solution Q5#
# Solution Q5 (full run + metrics + plots)
import torch, torch.nn as nn
from torch.utils.data import Dataset, DataLoader
# Data
df_mp = df[["MolWt","LogP","TPSA","NumRings","Melting Point"]].dropna().copy()
X = df_mp[["MolWt","LogP","TPSA","NumRings"]].values.astype(np.float32)
y = df_mp["Melting Point"].values.astype(np.float32).reshape(-1,1)
Xtr, Xte, ytr, yte = train_test_split(X, y, test_size=0.2, random_state=15)
scaler = StandardScaler().fit(Xtr)
Xtr_s = scaler.transform(Xtr).astype(np.float32)
Xte_s = scaler.transform(Xte).astype(np.float32)
class NumpyDataset(Dataset):
def __init__(self, X, y):
self.X = torch.from_numpy(X)
self.y = torch.from_numpy(y)
def __len__(self): return len(self.X)
def __getitem__(self, i): return self.X[i], self.y[i]
train_loader = DataLoader(NumpyDataset(Xtr_s, ytr), batch_size=64, shuffle=True)
in_dim = Xtr_s.shape[1]
model = nn.Sequential(
nn.Linear(in_dim, 32), nn.ReLU(),
nn.Linear(32, 16), nn.ReLU(),
nn.Linear(16, 1)
)
loss_fn = nn.MSELoss()
opt = torch.optim.Adam(model.parameters(), lr=1e-3, weight_decay=1e-3)
train_losses = []
model.train()
for epoch in range(200):
batch_losses = []
for xb, yb in train_loader:
pred = model(xb)
loss = loss_fn(pred, yb)
opt.zero_grad(); loss.backward(); opt.step()
batch_losses.append(loss.item())
train_losses.append(np.mean(batch_losses))
# Evaluate
model.eval()
with torch.no_grad():
yhat = model(torch.from_numpy(Xte_s)).numpy()
print(f"MSE: {mean_squared_error(yte, yhat):.3f}")
print(f"MAE: {mean_absolute_error(yte, yhat):.3f}")
print(f"R2: {r2_score(yte, yhat):.3f}")
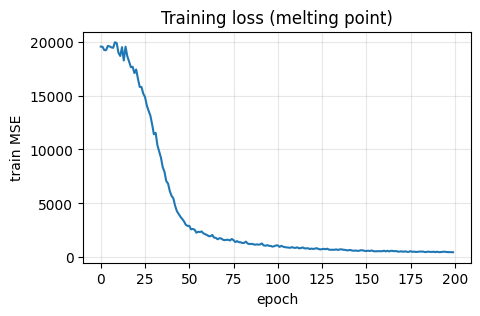
# Learning curve
plt.figure(figsize=(5,3))
plt.plot(train_losses)
plt.xlabel("epoch"); plt.ylabel("train MSE"); plt.title("Training loss (melting point)")
plt.grid(alpha=0.3)
plt.show()
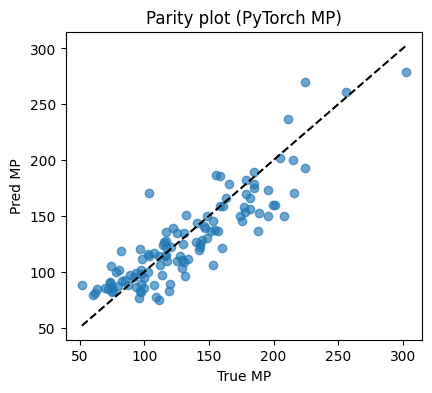
# Parity plot
plt.figure(figsize=(4.6,4))
plt.scatter(yte, yhat, alpha=0.65)
lims = [min(yte.min(), yhat.min()), max(yte.max(), yhat.max())]
plt.plot(lims, lims, "k--")
plt.xlabel("True MP"); plt.ylabel("Pred MP"); plt.title("Parity plot (PyTorch MP)")
plt.show()
MSE: 490.851
MAE: 17.622
R2: 0.769