Lecture 4 - Chemical Structure Identifier#
Learning goals#
Understand PubChem as a data service via PUG-REST and PUG-View.
Form URLs that return JSON, TXT or images.
Resolve names, SMILES and CAS numbers to CIDs.
Retrieve IUPAC, SMILES, InChIKey and selected properties.
Use the NCI Chemical Identifier Resolver (CIR) as a second query path.
Build small helper functions with error handling and fallbacks.
Practice with a real Excel sheet of ligands that contains CAS numbers.
Create a tiny widget that accepts name or SMILES or CAS and returns a summary and a GIF.
1. PubChem PUG-REST Basics#
1.1 Setup#
1.2 Background Information#
Instead of going to PubChem’s main site and manually searching for a compound, we can also use a direct URL to query PubChem’s REST API. This allows us to send structured requests and retrieve data in machine-readable formats such as JSON, XML, or plain text.
Other latest features of PubChem can be found in Nucleic Acids Research, 2025, 53 (D1), D1516–D1525.
The figure below illustrates the general workflow of PUG-REST: provide an input (like a compound name), choose an operation (for example, retrieving a CID), and specify the output format. Using URLs in this way not only automates lookups but also integrates PubChem data smoothly into code and analysis.
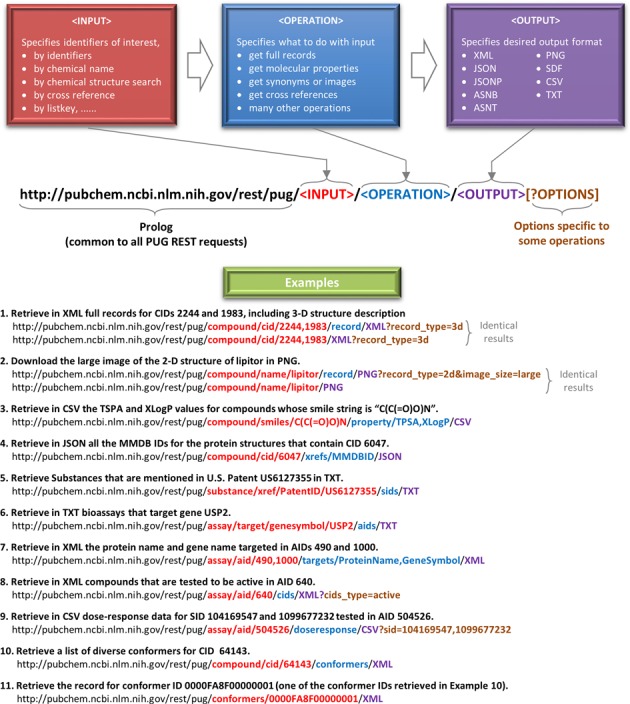
Here is a summary:
Input: name, CID, InChIKey, SMILES, or CAS number.
Operation: what property you want (CID, IUPAC, SMILES, etc.).
Output: JSON, XML, or TXT.
Here are some example URLs you can click and explore directly in a browser:
https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/name/aspirin/property/SMILES/TXT: Returns the SMILES string for aspirin.
https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/cid/2244/property/SMILES/JSON: Returns the SMILES string for a compound with PubChem CID 2244.
https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/name/aspirin/cids/JSON: Returns the PubChem Compound ID (CID) for aspirin.
https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/cid/2244/property/IUPACName/JSON: Returns the standardized IUPAC name.
https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/cid/2244/property/CanonicalSMILES/JSON: Returns the canonical SMILES string representation.
For more information on how to use PUG-REST URL, please see
Note
The key idea is that PubChem is not only a website but also a programmatic data service. A well-formed URL acts like a query to their database.
If you are interested in learning more about PubChem URL, please read:
Note
Always read the URL as:
Base:
https://pubchem.ncbi.nlm.nih.gov/rest/pugSection:
compound(since we work with compounds)Identifier type:
name,cid,smiles,casAction: what property to fetch
Format:
JSON,XML,TXT
2. From name to CID and properties#
2.1 Form the URL#
name = "acetaminophen"
url = f"https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/name/{quote_plus(name)}/cids/JSON"
print(url)
https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/name/acetaminophen/cids/JSON
Note
Try pasting this URL into your browser. You should see a JSON object with "CID".
2.2 Request and parse#
# Install requests if you do not have it
try:
import requests
except Exception:
%pip -q install requests
import requests
from urllib.parse import quote_plus
r = requests.get(url, timeout=30)
data = r.json()
print(data)
{'IdentifierList': {'CID': [1983]}}
Break it down:
print(data.keys())
print(data.get("IdentifierList", {}))
dict_keys(['IdentifierList'])
{'CID': [1983]}
cid_list = data.get("IdentifierList", {}).get("CID", [])
cid = cid_list[0]
print("CID:", cid)
CID: 1983
2.3 IUPAC name#
cid = 2244 # aspirin
url = f"https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/cid/{cid}/property/IUPACName/JSON"
r = requests.get(url, timeout=30)
r.raise_for_status()
props = r.json()["PropertyTable"]["Properties"][0]
print("IUPAC:", props["IUPACName"])
IUPAC: 2-acetyloxybenzoic acid
2.4 Canonical vs isomeric SMILES#
canon = requests.get(f"https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/cid/{cid}/property/CanonicalSMILES/TXT", timeout=30).text.strip()
iso = requests.get(f"https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/cid/{cid}/property/IsomericSMILES/TXT", timeout=30).text.strip()
print("Canonical:", canon)
print("Isomeric :", iso)
Canonical: CC(=O)OC1=CC=CC=C1C(=O)O
Isomeric : CC(=O)OC1=CC=CC=C1C(=O)O
Note
Canonical SMILES is normalized and does not carry stereochemistry.
Isomeric SMILES includes stereochemistry and isotopes if available.
2.5 Safer helpers#
def safe_get_json(url, timeout=30):
r = requests.get(url, timeout=timeout)
r.raise_for_status()
return r.json()
def safe_get_text(url, timeout=30):
r = requests.get(url, timeout=timeout)
r.raise_for_status()
return r.text.strip()
def safe_get_bytes(url, timeout=30):
r = requests.get(url, timeout=timeout)
r.raise_for_status()
return r.content
2.6 Write convenience functions (by name)#
def pubchem_by_name(name, isomeric=True):
enc = quote_plus(name.strip())
kind = "IsomericSMILES" if isomeric else "CanonicalSMILES"
smi_url = f"https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/name/{enc}/property/{kind}/TXT"
smiles = safe_get_text(smi_url)
cid_url = f"https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/name/{enc}/cids/JSON"
cid_list = safe_get_json(cid_url).get("IdentifierList", {}).get("CID", [])
if not cid_list:
raise ValueError("No CID found")
cid = cid_list[0]
iupac_url = f"https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/cid/{cid}/property/IUPACName/TXT"
iupac = safe_get_text(iupac_url)
inchikey_url = f"https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/cid/{cid}/property/InChIKey/TXT"
inchikey = safe_get_text(inchikey_url)
return {"input": name, "cid": cid, "smiles": smiles, "iupac": iupac, "inchikey": inchikey}
pubchem_by_name("ibuprofen")
{'input': 'ibuprofen',
'cid': 3672,
'smiles': 'CC(C)CC1=CC=C(C=C1)C(C)C(=O)O',
'iupac': '2-[4-(2-methylpropyl)phenyl]propanoic acid',
'inchikey': 'HEFNNWSXXWATRW-UHFFFAOYSA-N'}
2.7 Write convenience functions (by CID)#
def pubchem_by_cid(cid, isomeric=True):
cid = int(cid)
kind = "IsomericSMILES" if isomeric else "CanonicalSMILES"
smi_url = f"https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/cid/{cid}/property/{kind}/TXT"
smiles = safe_get_text(smi_url)
iupac_url = f"https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/cid/{cid}/property/IUPACName/TXT"
iupac = safe_get_text(iupac_url)
inchikey_url = f"https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/cid/{cid}/property/InChIKey/TXT"
inchikey = safe_get_text(inchikey_url)
return {"cid": cid, "smiles": smiles, "iupac": iupac, "inchikey": inchikey}
pubchem_by_cid(2244)
{'cid': 2244,
'smiles': 'CC(=O)OC1=CC=CC=C1C(=O)O',
'iupac': '2-acetyloxybenzoic acid',
'inchikey': 'BSYNRYMUTXBXSQ-UHFFFAOYSA-N'}
3. Other PubChem features#
3.1 Selected computed properties#
def pubchem_properties(cid, fields=("MolecularWeight","XLogP","ExactMass","HBondDonorCount","HBondAcceptorCount","TPSA")):
cid = int(cid)
field_str = ",".join(fields)
url = f"https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/cid/{cid}/property/{field_str}/JSON"
data = safe_get_json(url)
return data["PropertyTable"]["Properties"][0]
pubchem_properties(2244)
{'CID': 2244,
'MolecularWeight': '180.16',
'XLogP': 1.2,
'ExactMass': '180.04225873',
'TPSA': 63.6,
'HBondDonorCount': 1,
'HBondAcceptorCount': 4}
3.2 Synonyms#
def pubchem_synonyms(cid, limit=10):
url = f"https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/cid/{int(cid)}/synonyms/JSON"
syns = safe_get_json(url).get("InformationList", {}).get("Information", [{}])[0].get("Synonym", [])
return syns[:limit]
pubchem_synonyms(2244, limit=8)
['aspirin',
'ACETYLSALICYLIC ACID',
'50-78-2',
'2-Acetoxybenzoic acid',
'2-(Acetyloxy)benzoic acid',
'O-Acetylsalicylic acid',
'o-Acetoxybenzoic acid',
'Acylpyrin']
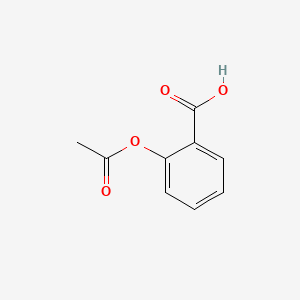
3.3 PNG depiction from PubChem#
def pubchem_png_by_cid(cid, size=300):
url = f"https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/cid/{int(cid)}/PNG?image_size={int(size)}x{int(size)}"
return safe_get_bytes(url)
png_bytes = pubchem_png_by_cid(2244, size=300)
display(Image(data=png_bytes))
3.4 PUG-View for safety snippets#
PUG-View serves formatted record content such as GHS classification. The path is:
https://pubchem.ncbi.nlm.nih.gov/rest/pug_view/data/compound/<cid>/JSON
The JSON is nested. The simple utility below searches for any block whose title contains a keyword.
def pug_view_find_sections(cid, keyword="GHS"):
url = f"https://pubchem.ncbi.nlm.nih.gov/rest/pug_view/data/compound/{int(cid)}/JSON"
data = safe_get_json(url)
hits = []
def walk(sections, path=""):
for sec in sections or []:
title = sec.get("TOCHeading", "") or sec.get("Title", "")
if keyword.lower() in title.lower():
hits.append({"title": title, "path": path + " > " + title, "section": sec})
walk(sec.get("Section"), path + " > " + title)
rec = data.get("Record", {})
walk(rec.get("Section"))
return hits
ghs_hits = pug_view_find_sections(2244, "GHS")
[x["title"] for x in ghs_hits][:5]
['GHS Classification', 'UN GHS Classification']
To show a short plain text summary, extract strings in the first hit:
def pug_view_text_from_hit(hit, max_lines=12):
lines = []
def walk(sec):
for i in sec.get("Information", []) or []:
sval = i.get("Value", {}).get("StringWithMarkup", [])
for s in sval:
txt = s.get("String", "").strip()
if txt:
lines.append(txt)
for ch in sec.get("Section", []) or []:
walk(ch)
walk(hit["section"])
return "\n".join(lines[:max_lines])
if ghs_hits:
print(pug_view_text_from_hit(ghs_hits[0]))
This chemical does not meet GHS hazard criteria for 0.3% (1 of 315) of reports.
Warning
H302 (95.6%): Harmful if swallowed [Warning Acute toxicity, oral]
H315 (20.6%): Causes skin irritation [Warning Skin corrosion/irritation]
H319 (22.2%): Causes serious eye irritation [Warning Serious eye damage/eye irritation]
H335 (20%): May cause respiratory irritation [Warning Specific target organ toxicity, single exposure; Respiratory tract irritation]
P261, P264, P264+P265, P270, P271, P280, P301+P317, P302+P352, P304+P340, P305+P351+P338, P319, P321, P330, P332+P317, P337+P317, P362+P364, P403+P233, P405, and P501
Aggregated GHS information provided per 315 reports by companies from 23 notifications to the ECHA C&L Inventory. Each notification may be associated with multiple companies.
Reported as not meeting GHS hazard criteria per 1 of 315 reports by companies.
There are 22 notifications provided by 314 of 315 reports by companies with hazard statement code(s).
Information may vary between notifications depending on impurities, additives, and other factors. The percentage value in parenthesis indicates the notified classification ratio from companies that provide hazard codes. Only hazard codes with percentage values above 10% are shown. For more detailed information, please visit ECHA C&L website.
Danger
4. NCI Chemical Identifier Resolver (CIR) as a second path#
Base URL:
https://cactus.nci.nih.gov/chemical/structure/<input>/<output>
Useful outputs: smiles, stdinchi, stdinchikey, names, cas, image, mw
def cir_get(q, out="smiles"):
q_enc = quote_plus(str(q).strip())
url = f"https://cactus.nci.nih.gov/chemical/structure/{q_enc}/{out}"
try:
if out == "image":
return safe_get_bytes(url)
else:
return safe_get_text(url)
except Exception as e:
return None
print("SMILES:", cir_get("aspirin", "smiles"), "...")
print("InChIKey:", cir_get("aspirin", "stdinchikey"))
print("CAS:", cir_get("aspirin", "cas"))
SMILES: CC(=O)Oc1ccccc1C(O)=O ...
InChIKey: InChIKey=BSYNRYMUTXBXSQ-UHFFFAOYSA-N
CAS: 50-78-2
11126-35-5
11126-37-7
2349-94-2
26914-13-6
98201-60-6
5. Mini widget#
Enter a name or CID or SMILES or CAS and show a quick summary. Generates a small GIF from a PNG depiction so it can be shared easily.
def quick_summary(query, source="PubChem"):
q = str(query).strip()
summary = {"query": q, "source": source}
try:
if source == "PubChem": # deafault PubChem
if q.isdigit():
info = pubchem_by_cid(int(q))
else:
info = pubchem_by_name(q)
summary.update(info)
png = pubchem_png_by_cid(summary["cid"], size=300)
summary["image"] = png
else: #use CIR
summary["smiles"] = cir_by_name(q, "smiles")
summary["inchikey"] = cir_by_name(q, "stdinchikey")
summary["iupac"] = cir_by_name(q, "iupac_name")
img = cir_by_name(q, "image")
summary["image"] = img if img else None
return summary
except Exception as e:
summary["error"] = str(e)
return summary
def run_fetch(b):
with out:
clear_output()
res = quick_summary(inp.value, source=src.value)
if "error" in res:
print("Error:", res["error"])
return
print("Source:", res.get("source"))
print("CID:", res.get("cid"))
print("IUPAC:", res.get("iupac"))
print("SMILES:", res.get("smiles"))
print("InChIKey:", res.get("inchikey"))
if res.get("image"):
display(Image(data=res["image"]))
else:
print("No image available")
if widgets is not None:
inp = widgets.Text(value="aspirin", description="Query:", layout=widgets.Layout(width="60%"))
src = widgets.ToggleButtons(options=["PubChem","CIR"], description="Source:")
btn = widgets.Button(description="Fetch", button_style="primary")
out = widgets.Output()
btn.on_click(run_fetch)
display(widgets.HBox([inp, src, btn]), out)
else:
print("ipywidgets is not available in this environment.")
6. Case study with CAS from Excel#
We will re-use the inventory file:
📂 organic ligands inventory.xlsx
https://raw.githubusercontent.com/zzhenglab/ai4chem/main/book/_data/organic_ligands_inventory.xlsx
import pandas as pd
url = "https://raw.githubusercontent.com/zzhenglab/ai4chem/main/book/_data/organic_ligands_inventory.xlsx"
df = pd.read_excel(url, engine="openpyxl")
df.head()
| CAS | Product Name | MW | Purity | Stock | |
|---|---|---|---|---|---|
| 0 | 2039-06-7 | 3,5-Diphenyl-4H-1,2,4-triazole | 221.263 | 98% | In Stock |
| 1 | 2127-07-3 | 1,2-Bis(2-(pyridin-4-yl)ethyl)disulfane | 276.420 | 98% | In Stock |
| 2 | 2359-09-3 | 5-tert-Butylisophthalic acid | 222.240 | 96% | In Stock |
| 3 | 2384-02-3 | 1H,1'H-3,3'-Bipyrazole | 134.140 | 98% | In Stock |
| 4 | 2790-09-2 | 4,6-Dimethylisophthalic acid | 194.186 | 98% | In Stock |
Why “openpyxl”?
In previous example, we directly read csv. However, to access excell spreedsheet, we need to specify engine="openpyxl".
6.1 Query PubChem by CAS with fallback#
def by_cas_with_fallback(cas, isomeric=True):
enc = quote_plus(str(cas))
try:
kind = "IsomericSMILES" if isomeric else "CanonicalSMILES"
smi_url = f"https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/name/{enc}/property/{kind}/TXT"
smiles = safe_get_text(smi_url)
cid_url = f"https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/name/{enc}/cids/TXT"
try:
cid = int(safe_get_text(cid_url).split()[0])
except Exception:
cid = None
except Exception:
smiles = cir_get(cas, "smiles")
cid = None
return {"cas": cas, "cid": cid, "smiles": smiles}
example_cas = df.loc[0, "CAS"]
by_cas_with_fallback(example_cas)
{'cas': '2039-06-7',
'cid': 16260,
'smiles': 'C1=CC=C(C=C1)C2=NC(=NN2)C3=CC=CC=C3'}
6.2 Enrich a small slice#
rows = []
for cas in df["CAS"].head(8):
try:
rec = by_cas_with_fallback(cas)
if rec["cid"]:
more = pubchem_properties(rec["cid"])
rec.update(more)
rows.append(rec)
except Exception:
rows.append({"cas": cas, "cid": None, "smiles": None})
pd.DataFrame(rows)
| cas | cid | smiles | CID | MolecularWeight | XLogP | ExactMass | TPSA | HBondDonorCount | HBondAcceptorCount | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2039-06-7 | 16260.0 | C1=CC=C(C=C1)C2=NC(=NN2)C3=CC=CC=C3 | 16260.0 | 221.26 | 3.3 | 221.095297364 | 41.6 | 1.0 | 2.0 |
| 1 | 2127-07-3 | 417543.0 | C1=CN=CC=C1CCSSCCC2=CC=NC=C2 | 417543.0 | 276.4 | 2.7 | 276.07549087 | 76.4 | 0.0 | 4.0 |
| 2 | 2359-09-3 | 75379.0 | CC(C)(C)C1=CC(=CC(=C1)C(=O)O)C(=O)O | 75379.0 | 222.24 | 2.6 | 222.08920892 | 74.6 | 2.0 | 4.0 |
| 3 | 2384-02-3 | 584077.0 | C1=C(NN=C1)C2=CC=NN2 | 584077.0 | 134.14 | 0.2 | 134.059246208 | 57.4 | 2.0 | 2.0 |
| 4 | 2790-09-2 | 314208.0 | CC1=CC(=C(C=C1C(=O)O)C(=O)O)C | 314208.0 | 194.18 | 1.7 | 194.05790880 | 74.6 | 2.0 | 4.0 |
| 5 | 2997-05-9 | NaN | None | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 6 | 3097-03-8 | 12262901.0 | C1=CC2=C(C=C1C3=CC4=C(C=C3)N=CN4)NC=N2 | 12262901.0 | 234.26 | 2.7 | 234.090546336 | 57.4 | 2.0 | 2.0 |
| 7 | 3711-03-3 | 13628397.0 | C1=CC2=C(C3=C1C4=C(C=C3)C(=O)OC4=O)C(=O)OC2=O | 13628397.0 | 268.18 | 1.9 | 268.00078784 | 86.7 | 0.0 | 6.0 |
7. Glossary#
- PUG-REST#
The PubChem REST interface that returns properties, identifiers and files via URL patterns.
- PUG-View#
A service that returns human-oriented record sections such as safety and vendor data in JSON.
- CID#
PubChem Compound ID for a unique compound record.
- SID#
PubChem Substance ID which refers to a submitted substance record.
- AID#
PubChem Assay ID for a bioassay record.
- SMILES#
Text line notation for molecules. Canonical SMILES omits stereochemistry. Isomeric SMILES includes it.
- InChI and InChIKey#
IUPAC identifiers. InChIKey is a hashed, fixed-length form that is easy to compare.
- GHS#
Globally Harmonized System of Classification and Labelling of Chemicals. PubChem PUG-View can include these sections.
- CIR#
NCI Chemical Identifier Resolver. A second service that converts between names, SMILES, InChI, InChIKey and more.
- sanitize#
RDKit process that checks valence, aromaticity and stereochemistry.
- descriptor#
Computed molecular property such as molecular weight or LogP.
8. Quick reference#
PubChem endpoints to remember
Name to CID:
/compound/name/<name>/cids/JSONCID to properties:
/compound/cid/<cid>/property/<fields>/JSONCID to PNG depiction:
/compound/cid/<cid>/PNG?image_size=300x300Synonyms:
/compound/cid/<cid>/synonyms/JSONPUG-View record:
/rest/pug_view/data/compound/<cid>/JSON
CIR endpoints to remember
/<input>/smiles,/<input>/stdinchi,/<input>/stdinchikey/<input>/namesand/<input>/casfor synonyms and CAS when available/<input>/imageto get a 2D depiction/<input>/mwto get molecular weight
9. In-class activity#
Each task is aligned with what we covered above. Work in pairs. Fill in the ... lines. Solutions are in Section 13.
9.1 Verify PubChem vs CIR agreement#
Given targets = ["caffeine", "ibuprofen", "acetaminophen"]
Get InChIKey from PubChem and from CIR.
Report whether they match.
targets = ["caffeine", "ibuprofen", "acetaminophen"]
for t in targets:
pc_key = ... #TO DO
cir_key = ... #TO DO
print(f"{t:14s} PubChem={pc_key} CIR={cir_key} match={pc_key==cir_key}")
9.2 Small properties table from names#
Use names ["caffeine", "acetaminophen", "ibuprofen"]. For each, fetch SMILES from PubChem, then compute MolWt, LogP, HBD, HBA, and TPSA.
#TO DO
9.3 Get a quick GHS note from PUG-View#
For CID 2244 retrieve GHS section text.
Print the first 8 lines.
#TO DO
9.4 Build a one-liner resolver#
Input list
["446157", "2244", "CCO", "482752", "c1ccccc1"]If it is digits treat as CID else try SMILES else treat as name.
Print
input=# CID=# SMILES=...and draw if RDKit is available.
items = ["446157", "2244", "CCO", "482752", "c1ccccc1"]
for q in items:
if ...
...#TO DO
print(f"input={...} CID={...} SMILES={...}")
9.5 Process dataset#
Load dataset “https://raw.githubusercontent.com/zzhenglab/ai4chem/main/book/_data/C_H_oxidation_dataset.csv”.
Examine the dataset structure using df.head() and select the SMILES strings from first 5 rows to query CIR (see section 4) to get number of freely rotatable bonds.
#TO DO
10. Solutions#
Solution 9.1#
targets = ["caffeine", "ibuprofen", "acetaminophen"]
for t in targets:
pc_key = pubchem_by_name(t)["inchikey"]
cir_key = cir_get(t, "stdinchikey")
print(f"{t:14s} PubChem={pc_key} CIR={cir_key} match={pc_key==cir_key}")
caffeine PubChem=RYYVLZVUVIJVGH-UHFFFAOYSA-N CIR=InChIKey=RYYVLZVUVIJVGH-UHFFFAOYSA-N match=False
ibuprofen PubChem=HEFNNWSXXWATRW-UHFFFAOYSA-N CIR=InChIKey=HEFNNWSXXWATRW-UHFFFAOYSA-N match=False
acetaminophen PubChem=RZVAJINKPMORJF-UHFFFAOYSA-N CIR=InChIKey=RZVAJINKPMORJF-UHFFFAOYSA-N match=False
Solution 9.2#
if Chem is not None:
import pandas as pd
rows = []
names = ["caffeine", "acetaminophen", "ibuprofen"]
for nm in names:
info = pubchem_by_name(nm)
smi = info["smiles"]
m = Chem.MolFromSmiles(smi)
rows.append({
"name": nm,
"smiles": smi,
"MolWt": Descriptors.MolWt(m),
"LogP": Crippen.MolLogP(m),
"HBD": rdMolDescriptors.CalcNumHBD(m),
"HBA": rdMolDescriptors.CalcNumHBA(m),
"TPSA": rdMolDescriptors.CalcTPSA(m)
})
pd.DataFrame(rows)
else:
print("RDKit is not available here.")
---------------------------------------------------------------------------
HTTPError Traceback (most recent call last)
Cell In[23], line 6
4 names = ["caffeine", "acetaminophen", "ibuprofen"]
5 for nm in names:
----> 6 info = pubchem_by_name(nm)
7 smi = info["smiles"]
8 m = Chem.MolFromSmiles(smi)
Cell In[10], line 8, in pubchem_by_name(name, isomeric)
5 smiles = safe_get_text(smi_url)
7 cid_url = f"https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/name/{enc}/cids/JSON"
----> 8 cid_list = safe_get_json(cid_url).get("IdentifierList", {}).get("CID", [])
9 if not cid_list:
10 raise ValueError("No CID found")
Cell In[9], line 3, in safe_get_json(url, timeout)
1 def safe_get_json(url, timeout=30):
2 r = requests.get(url, timeout=timeout)
----> 3 r.raise_for_status()
4 return r.json()
File ~\AppData\Local\Programs\Python\Python313\Lib\site-packages\requests\models.py:1026, in Response.raise_for_status(self)
1021 http_error_msg = (
1022 f"{self.status_code} Server Error: {reason} for url: {self.url}"
1023 )
1025 if http_error_msg:
-> 1026 raise HTTPError(http_error_msg, response=self)
HTTPError: 503 Server Error: PUGREST.ServerBusy for url: https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/name/ibuprofen/cids/JSON
Solution 9.3#
hits = pug_view_find_sections(2244, "GHS")
print(pug_view_text_from_hit(hits[0], max_lines=8))
Solution 9.4#
if Chem is not None:
find = Chem.MolFromSmiles("Cl")
put = Chem.MolFromSmiles("F")
mol = Chem.MolFromSmiles("Clc1ccc(cc1)C(=O)O")
out = Chem.ReplaceSubstructs(mol, find, put, replaceAll=True)[0]
print(Chem.MolToSmiles(out))
Draw.MolToImage(out, size=(300, 220))
else:
print("RDKit is not available here.")
Solution 9.5#
items = ["446157", "2244", "CCO", "482752", "c1ccccc1"]
for q in items:
if q.isdigit():
info = pubchem_by_cid(int(q))
smi, cid = info["smiles"], info["cid"]
elif Chem is not None and Chem.MolFromSmiles(q):
smi, cid = q, None
else:
info = pubchem_by_name(q)
smi, cid = info["smiles"], info["cid"]
print(f"input={q} CID={cid} SMILES={smi}")
if Chem is not None:
m = Chem.MolFromSmiles(smi)
display(Draw.MolToImage(m, size=(220, 180)))



